

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.14
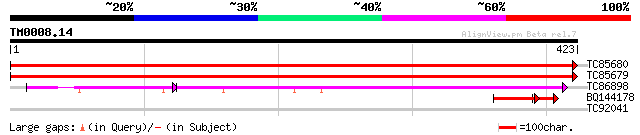
(423 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC85680 homologue to GP|15919089|emb|CAC86996. ATP citrate lyase... 806 0.0
TC85679 homologue to GP|15919089|emb|CAC86996. ATP citrate lyase... 774 0.0
TC86898 similar to GP|18376007|emb|CAB91741. probable ATP citrat... 241 2e-79
BQ144178 similar to GP|16648642|gb| ATP-citrate lyase subunit A ... 51 2e-09
TC92041 similar to GP|1262234|gb|AAB48690.1| RNA polymerase {Hum... 28 6.7
>TC85680 homologue to GP|15919089|emb|CAC86996. ATP citrate lyase b-subunit
{Lupinus albus}, complete
Length = 1698
Score = 806 bits (2083), Expect = 0.0
Identities = 402/423 (95%), Positives = 415/423 (98%)
Frame = +1
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
MARKKIREYDSKRL+KEHFKRLSGK+LPIKSAQVT +TDF+ELQDKE WLSSSKLVVKPD
Sbjct: 169 MARKKIREYDSKRLLKEHFKRLSGKDLPIKSAQVTEATDFTELQDKEQWLSSSKLVVKPD 348
Query: 61 MLFGKRGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 120
MLFGKRGKSGLVALNLD AQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEE+YLN
Sbjct: 349 MLFGKRGKSGLVALNLDLAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEYYLN 528
Query: 121 IVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVATLPLEIKGEIEE 180
IVS+RLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEI+APLVATLPLEIKGEIEE
Sbjct: 529 IVSDRLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIIAPLVATLPLEIKGEIEE 708
Query: 181 FLKVIFTLFQDLDFTFLEMNPFTLVDGKPCPLDMRGELDDTAAFKNFKKWGNIEFPLPFG 240
FLKVIFTLFQDLDFTFLEMNPFTLVDGKP PLDMRGELDDTAAFKNFKKWGNIEFPLPFG
Sbjct: 709 FLKVIFTLFQDLDFTFLEMNPFTLVDGKPYPLDMRGELDDTAAFKNFKKWGNIEFPLPFG 888
Query: 241 RVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGN 300
RVMSATESF+H LDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFA+ELGN
Sbjct: 889 RVMSATESFIHGLDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFANELGN 1068
Query: 301 YAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE 360
YAEYSGAP E+EVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE
Sbjct: 1069YAEYSGAPNEEEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE 1248
Query: 361 KESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATMTGICKEAIQCIT 420
KESKLKAARMH+YVRRGGPNYQKGL MRALGEEIGIPIEVYGPEATMTGICK+AIQCIT
Sbjct: 1249KESKLKAARMHLYVRRGGPNYQKGLEKMRALGEEIGIPIEVYGPEATMTGICKQAIQCIT 1428
Query: 421 AAA 423
A+A
Sbjct: 1429ASA 1437
>TC85679 homologue to GP|15919089|emb|CAC86996. ATP citrate lyase b-subunit
{Lupinus albus}, complete
Length = 1732
Score = 774 bits (1998), Expect = 0.0
Identities = 383/423 (90%), Positives = 403/423 (94%)
Frame = +2
Query: 1 MARKKIREYDSKRLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWLSSSKLVVKPD 60
MARKKIREYDSKRL+KEHFKR+SG++LPIKSAQVT STD +EL +KEPWLSSSKLVVKPD
Sbjct: 83 MARKKIREYDSKRLLKEHFKRISGQDLPIKSAQVTESTDINELVEKEPWLSSSKLVVKPD 262
Query: 61 MLFGKRGKSGLVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 120
MLFGKRGKSGLVALNLD A VA+FVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN
Sbjct: 263 MLFGKRGKSGLVALNLDLAGVAAFVKERLGKEVEMGGCKGPITTFIVEPFIPHNEEFYLN 442
Query: 121 IVSERLGNSISFSECGGIDIEENWDKVKTVFIPTGVSLTSEIVAPLVATLPLEIKGEIEE 180
IVS+RLGNSISFSECGGIDIEENWDKVKTVFIPTG SLT+E ++PL+ATLPLEIKGE+E+
Sbjct: 443 IVSDRLGNSISFSECGGIDIEENWDKVKTVFIPTGESLTAENISPLIATLPLEIKGELED 622
Query: 181 FLKVIFTLFQDLDFTFLEMNPFTLVDGKPCPLDMRGELDDTAAFKNFKKWGNIEFPLPFG 240
FLKVIF +FQDLDFTFLEMNPF LVDGKP PLDMRGELDDTA FKNFKKWGNIEFPLPFG
Sbjct: 623 FLKVIFNIFQDLDFTFLEMNPFALVDGKPYPLDMRGELDDTATFKNFKKWGNIEFPLPFG 802
Query: 241 RVMSATESFVHELDEKTSASLKFTVLNPKGRIWTMVAGGGASVIYADTVGDLGFASELGN 300
RVMS TESF+H LDEKTSASLKFTVLNP GRIWTMVAGGGASVIYADTVGDLG+A ELGN
Sbjct: 803 RVMSPTESFIHGLDEKTSASLKFTVLNPVGRIWTMVAGGGASVIYADTVGDLGYAPELGN 982
Query: 301 YAEYSGAPKEDEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE 360
YAEYSGAPKE EVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE
Sbjct: 983 YAEYSGAPKEGEVLQYARVVIDCATANPDGQKRALVIGGGIANFTDVAATFSGIIRALKE 1162
Query: 361 KESKLKAARMHIYVRRGGPNYQKGLALMRALGEEIGIPIEVYGPEATMTGICKEAIQCIT 420
KE KLK A MHIYVRRGGPNYQ+GLA MRALGEEIGIPIEVYGPEATMTGICK+AIQCIT
Sbjct: 1163KEQKLKEANMHIYVRRGGPNYQRGLAKMRALGEEIGIPIEVYGPEATMTGICKQAIQCIT 1342
Query: 421 AAA 423
A+A
Sbjct: 1343ASA 1351
>TC86898 similar to GP|18376007|emb|CAB91741. probable ATP citrate lyase
subunit 2 {Neurospora crassa}, complete
Length = 2389
Score = 241 bits (614), Expect(2) = 2e-79
Identities = 132/329 (40%), Positives = 191/329 (57%), Gaps = 37/329 (11%)
Frame = +2
Query: 125 RLGNSISFSECGGIDIEENWDKVKTVFIPTGVSL---TSEIVAPLVATLPLEIKGEIEEF 181
R G+ I F+ GG+D+ + K + + IP +S EI A L+ +P + + +F
Sbjct: 1013 RDGDWILFTHEGGVDVGDVDAKAEKLLIPVDLSQYPSNEEIAATLLKKVPKGVHNVLVDF 1192
Query: 182 LKVIFTLFQDLDFTFLEMNPFTLVDGKPCP------LDMRGELDDTAAFKNFKKWG---- 231
+ ++ ++ D FT+LE+NP ++ + LD+ +LD TA F+ KW
Sbjct: 1193 ISRLYAVYVDCQFTYLEINPLVVIPNEDATSASVHFLDLAAKLDQTADFECGVKWAIARS 1372
Query: 232 -----------------------NIEFPLPFGRVMSATESFVHELDEKTSASLKFTVLNP 268
+EFP PFGR ++ E+++ +LD KT ASLK TVLNP
Sbjct: 1373 PAALGLTNVASSNGEKVNIDAGPPMEFPAPFGRELTKEEAYIADLDAKTGASLKLTVLNP 1552
Query: 269 KGRIWTMVAGGGASVIYADTVGDLGFASELGNYAEYSGAPKEDEVLQYARVVIDCATANP 328
GRIWT+VAGGGASV+YAD + GFA EL NY EYSGAP E + YAR V+D P
Sbjct: 1553 NGRIWTLVAGGGASVVYADAIASSGFADELANYGEYSGAPTESQTYHYARTVLDLMLRAP 1732
Query: 329 DGQK-RALVIGGGIANFTDVAATFSGIIRALKEKESKLKAARMHIYVRRGGPNYQKGLAL 387
+K + L IGGGIANFT+VA+TF G+IRAL++ +L + I+VRR GPNYQ+GL
Sbjct: 1733 LTEKGKVLFIGGGIANFTNVASTFKGVIRALRDYAKQLNEHKTQIWVRRAGPNYQEGLKN 1912
Query: 388 MRALGEEIGIPIEVYGPEATMTGICKEAI 416
M+A +E+G+ +++GPE ++GI A+
Sbjct: 1913 MKAATQELGLDAKIFGPEMHVSGIVPLAL 1999
Score = 73.2 bits (178), Expect(2) = 2e-79
Identities = 43/117 (36%), Positives = 65/117 (54%), Gaps = 4/117 (3%)
Frame = +1
Query: 13 RLVKEHFKRLSGKELPIKSAQVTGSTDFSELQDKEPWL--SSSKLVVKPDMLFGKRGKSG 70
RL HF + E + A+VT PW+ +K V KPD L +RGKSG
Sbjct: 697 RLCSLHFPEDANVEDILNQAEVTF-----------PWVLQPGAKFVAKPDQLIKRRGKSG 843
Query: 71 LVALNLDFAQVASFVKERLGKEVEMGGCKGPITTFIVEPFIPH--NEEFYLNIVSER 125
L+ALN + + +++ ER GKE ++ G + F+VEPF+PH + E+Y+NI+S +
Sbjct: 844 LLALNKTWPEAKAWIAERAGKEQKVEHTTGVLRQFLVEPFVPHPQDTEYYINIMSRQ 1014
>BQ144178 similar to GP|16648642|gb| ATP-citrate lyase subunit A {Arabidopsis
thaliana}, partial (7%)
Length = 657
Score = 50.8 bits (120), Expect(2) = 2e-09
Identities = 22/34 (64%), Positives = 27/34 (78%)
Frame = +2
Query: 362 ESKLKAARMHIYVRRGGPNYQKGLALMRALGEEI 395
+ LK + MHIYV+ GGPNYQKGL M+ALGE+I
Sbjct: 35 DQNLKESHMHIYVKIGGPNYQKGLTKMKALGEDI 136
Score = 28.9 bits (63), Expect(2) = 2e-09
Identities = 14/19 (73%), Positives = 14/19 (73%)
Frame = +3
Query: 391 LGEEIGIPIEVYGPEATMT 409
LG IPIEVYGP ATMT
Sbjct: 123 LGRTSCIPIEVYGP*ATMT 179
>TC92041 similar to GP|1262234|gb|AAB48690.1| RNA polymerase {Human
parainfluenza virus 3}, partial (0%)
Length = 606
Score = 28.1 bits (61), Expect = 6.7
Identities = 16/34 (47%), Positives = 21/34 (61%), Gaps = 1/34 (2%)
Frame = -1
Query: 7 REYDSKRL-VKEHFKRLSGKELPIKSAQVTGSTD 39
R+YD R V+E K+ G ELP AQV+GS +
Sbjct: 297 RDYDLNRSQVRELIKQYLGLELPDDKAQVSGSKE 196
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.138 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,121,166
Number of Sequences: 36976
Number of extensions: 136840
Number of successful extensions: 567
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 564
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 566
length of query: 423
length of database: 9,014,727
effective HSP length: 99
effective length of query: 324
effective length of database: 5,354,103
effective search space: 1734729372
effective search space used: 1734729372
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0008.14


