

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.13
(259 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
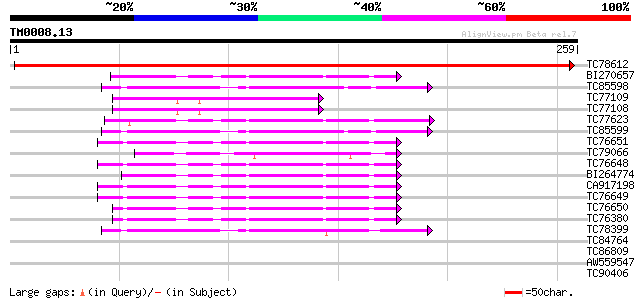
Score E
Sequences producing significant alignments: (bits) Value
TC78612 similar to GP|9759092|dbj|BAB09661.1 signal recognition ... 457 e-129
BI270657 homologue to PIR|T52341|T52 ADP-ribosylation factor [im... 47 6e-06
TC85598 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 46 2e-05
TC77109 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein... 45 4e-05
TC77108 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein... 45 4e-05
TC77623 homologue to GP|1654142|gb|AAB17725.1| small GTP-binding... 44 5e-05
TC85599 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 44 6e-05
TC76651 PIR|T52341|T52341 ADP-ribosylation factor [imported] - r... 43 1e-04
TC79066 homologue to PIR|T51561|T51561 ADP-ribosylation factor-l... 43 1e-04
TC76648 homologue to PIR|T52341|T52341 ADP-ribosylation factor [... 43 1e-04
BI264774 homologue to PIR|D49993|D49 ADP-ribosylation factor - A... 43 1e-04
CA917198 GP|20161472|db ADP-ribosylation factor {Oryza sativa (j... 43 1e-04
TC76649 homologue to GP|20161472|dbj|BAB90396. ADP-ribosylation ... 43 1e-04
TC76650 GP|965483|dbj|BAA08259.1| DcARF1 {Daucus carota}, complete 42 2e-04
TC76380 homologue to PIR|T52341|T52341 ADP-ribosylation factor [... 42 2e-04
TC78399 homologue to SP|O04834|SARA_ARATH GTP-binding protein SA... 42 2e-04
TC84764 similar to PIR|S71584|S71584 GTP-binding protein BG1 - A... 40 0.001
TC86809 homologue to PIR|S35701|S35701 translation elongation fa... 37 0.010
AW559547 weakly similar to GP|12654451|gb| ADP-ribosylation fact... 36 0.013
TC90406 similar to GP|11994726|dbj|BAB03042. contains similarity... 36 0.017
>TC78612 similar to GP|9759092|dbj|BAB09661.1 signal recognition particle
receptor beta subunit-like protein {Arabidopsis
thaliana}, partial (84%)
Length = 963
Score = 457 bits (1175), Expect = e-129
Identities = 224/256 (87%), Positives = 241/256 (93%)
Frame = +3
Query: 3 ELEHWKEELSRLWLTASDYLRQVPPEQLYIAAAIAVFTTLLLLSLRLFKRAKTNTVVLTG 62
ELE WKE+ S LW A+DYLR VPP QLY AAAIA+FTTLLLL LR+ KR K+NT+VLTG
Sbjct: 72 ELEQWKEQFSHLWNVANDYLRDVPPNQLYAAAAIAIFTTLLLLFLRVLKRTKSNTIVLTG 251
Query: 63 LTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPGHSRLR 122
L+GSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILH ET KKGK+KPVHIVDVPGHSRLR
Sbjct: 252 LSGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHGETAKKGKIKPVHIVDVPGHSRLR 431
Query: 123 PKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNKTDKVT 182
PKLDE+LPQAAG+VFVVDA+DFLPNCRAASEYLYD+LTKGSVV+KKIPLLILCNKTDKVT
Sbjct: 432 PKLDEFLPQAAGIVFVVDALDFLPNCRAASEYLYDLLTKGSVVRKKIPLLILCNKTDKVT 611
Query: 183 AHTKEFIRRQLEKEIDKLRASRSAVSDADVTNEFTLGVPGEAFSFTQCCNKVTTADASGL 242
AHTKEFIRRQ+EKEIDKLR+SRSAVS+ADVTNEFTLGVPGE FSFTQC NKVTTADASGL
Sbjct: 612 AHTKEFIRRQIEKEIDKLRSSRSAVSEADVTNEFTLGVPGEPFSFTQCSNKVTTADASGL 791
Query: 243 TGEISQLEEFIREYVK 258
TGEISQL+EFIREYVK
Sbjct: 792 TGEISQLQEFIREYVK 839
>BI270657 homologue to PIR|T52341|T52 ADP-ribosylation factor [imported] -
rice, partial (64%)
Length = 468
Score = 47.4 bits (111), Expect = 6e-06
Identities = 37/133 (27%), Positives = 63/133 (46%)
Frame = +3
Query: 47 LRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGK 106
+RLF K +++ GL +GKT + Y+L+ G + + P G + ET +
Sbjct: 63 MRLFYAKKEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKN 218
Query: 107 VKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVK 166
V + DV G ++RP Y G++FVVD+ D A + L+ +L++ +
Sbjct: 219 VS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RERILEARDELHRMLSEDEL-- 386
Query: 167 KKIPLLILCNKTD 179
+ LL+ NK D
Sbjct: 387 RDATLLVFANKQD 425
>TC85598 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 900
Score = 45.8 bits (107), Expect = 2e-05
Identities = 45/151 (29%), Positives = 69/151 (44%)
Frame = +3
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 201 VLASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQYPTSEELSI------ 356
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y Q VV++VDA D + E L +L+
Sbjct: 357 --GKIK-FKAFDLGGHQIARRVWKDYYAQVDAVVYLVDAYDKERFAESKKE-LDALLSDE 524
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
S+ +P L+L NK D A +++ +R L
Sbjct: 525 SLA--NVPFLVLGNKIDIPYAASEDELRHHL 611
>TC77109 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein typA
(tyrosine phosphorylated protein A) {Arabidopsis
thaliana}, partial (27%)
Length = 672
Score = 44.7 bits (104), Expect = 4e-05
Identities = 29/106 (27%), Positives = 47/106 (43%), Gaps = 10/106 (9%)
Frame = +3
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQL-------RDGSTHQGTV---TSMEPNEGTFIL 97
+L +RA + + GKT L + RD T Q + +E G IL
Sbjct: 165 KLMRRADIRNIAIVAHVDHGKTTLVDAMLKQTKVFRDNQTVQERIMDSNDLERERGITIL 344
Query: 98 HSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVD 143
T+ K ++I+D PGHS +++ L G++ VVD+V+
Sbjct: 345 SKNTSVTYKDAKINIIDTPGHSDFGGEVERILNMVEGILLVVDSVE 482
>TC77108 similar to GP|9758030|dbj|BAB08691.1 GTP-binding protein typA
(tyrosine phosphorylated protein A) {Arabidopsis
thaliana}, partial (98%)
Length = 2589
Score = 44.7 bits (104), Expect = 4e-05
Identities = 29/106 (27%), Positives = 47/106 (43%), Gaps = 10/106 (9%)
Frame = +3
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQL-------RDGSTHQGTV---TSMEPNEGTFIL 97
+L +RA + + GKT L + RD T Q + +E G IL
Sbjct: 375 KLMRRADIRNIAIVAHVDHGKTTLVDAMLKQTKVFRDNQTVQERIMDSNDLERERGITIL 554
Query: 98 HSETTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVD 143
T+ K ++I+D PGHS +++ L G++ VVD+V+
Sbjct: 555 SKNTSVTYKDAKINIIDTPGHSDFGGEVERILNMVEGILLVVDSVE 692
>TC77623 homologue to GP|1654142|gb|AAB17725.1| small GTP-binding protein
ARF {Brassica rapa}, complete
Length = 1195
Score = 44.3 bits (103), Expect = 5e-05
Identities = 43/154 (27%), Positives = 67/154 (42%), Gaps = 3/154 (1%)
Frame = +1
Query: 44 LLSLRLFKRA---KTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
L+ RLF K +++ GL +GKT + Y+L+ G V S P G + E
Sbjct: 271 LVFTRLFSSVFGNKEARILVLGLDNAGKTTILYRLQ-----MGEVVSTIPTIG---FNVE 426
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + +K + D+ G + +RP Y P +++VVD+ D A E + IL
Sbjct: 427 TVQYNNIK-FQVWDLGGQTSIRPYWRCYFPNTQAIIYVVDSSD-TDRLVIAREEFHAILE 600
Query: 161 KGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQLE 194
+ + K +LI NK D A + LE
Sbjct: 601 EEEL--KGAVVLIFANKQDLPGALDDAAVTESLE 696
>TC85599 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 999
Score = 43.9 bits (102), Expect = 6e-05
Identities = 45/151 (29%), Positives = 69/151 (44%)
Frame = +2
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L SL L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 182 ILASLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQHPTSEELSI------ 337
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKG 162
GK+K D+ GH R +Y + VV++VDA D + E L +L+
Sbjct: 338 --GKIK-FKAFDLGGHQIARRVWKDYYAKVDAVVYLVDAYDKERFAESKKE-LDALLSDE 505
Query: 163 SVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
S+ +P LIL NK D A +++ +R L
Sbjct: 506 SLA--NVPFLILGNKIDIPYAASEDELRYHL 592
>TC76651 PIR|T52341|T52341 ADP-ribosylation factor [imported] - rice,
complete
Length = 1044
Score = 43.1 bits (100), Expect = 1e-04
Identities = 38/139 (27%), Positives = 64/139 (45%)
Frame = +2
Query: 41 TLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
T L RLF + K +++ GL +GKT + Y+L+ G + + P G + E
Sbjct: 122 TFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVE 274
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + + + DV G ++RP Y G++FVVD+ D A + L+ +L
Sbjct: 275 TVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLN 448
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + + LL+ NK D
Sbjct: 449 EDEL--RDAVLLVFANKQD 499
>TC79066 homologue to PIR|T51561|T51561 ADP-ribosylation factor-like protein
- Arabidopsis thaliana, partial (95%)
Length = 839
Score = 43.1 bits (100), Expect = 1e-04
Identities = 39/127 (30%), Positives = 58/127 (44%), Gaps = 5/127 (3%)
Frame = +2
Query: 58 VVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPV--HIVDV 115
VV+ GL +GKT + Y+L H G V S P G + +K + K V + DV
Sbjct: 137 VVMLGLDAAGKTTILYKL-----HIGEVLSTVPTIGFNV------EKVQYKNVVFTVWDV 283
Query: 116 PGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEY---LYDILTKGSVVKKKIPLL 172
G +LRP Y G+++VVD++D +A E+ + D SV+ L
Sbjct: 284 GGQEKLRPLWRHYFNNTDGLIYVVDSLDRERISQAKQEFQAIINDPFMLNSVI------L 445
Query: 173 ILCNKTD 179
+ NK D
Sbjct: 446 VFANKQD 466
>TC76648 homologue to PIR|T52341|T52341 ADP-ribosylation factor [imported] -
rice, complete
Length = 1357
Score = 43.1 bits (100), Expect = 1e-04
Identities = 38/139 (27%), Positives = 64/139 (45%)
Frame = +3
Query: 41 TLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
T L RLF + K +++ GL +GKT + Y+L+ G + + P G + E
Sbjct: 519 TFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVE 671
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + + + DV G ++RP Y G++FVVD+ D A + L+ +L
Sbjct: 672 TVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLN 845
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + + LL+ NK D
Sbjct: 846 EDEL--RDAVLLVFANKQD 896
>BI264774 homologue to PIR|D49993|D49 ADP-ribosylation factor - Ajellomyces
capsulata, partial (91%)
Length = 665
Score = 43.1 bits (100), Expect = 1e-04
Identities = 35/128 (27%), Positives = 59/128 (45%)
Frame = +3
Query: 52 RAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVH 111
R K +++ GL +GKT + Y+L+ G + + P G + ET + ++
Sbjct: 6 RKKEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNIQ-FT 158
Query: 112 IVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPL 171
+ DV G ++RP Y G++FVVD+ D A E L +L + + + L
Sbjct: 159 VWDVGGQDKIRPLWRHYFQNTQGIIFVVDSND-RDRIVEAREELQRMLNEDEL--RDAIL 329
Query: 172 LILCNKTD 179
L+ NK D
Sbjct: 330 LVFANKQD 353
>CA917198 GP|20161472|db ADP-ribosylation factor {Oryza sativa (japonica
cultivar-group)}, partial (43%)
Length = 780
Score = 43.1 bits (100), Expect = 1e-04
Identities = 38/139 (27%), Positives = 64/139 (45%)
Frame = +3
Query: 41 TLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
T L RLF + K +++ GL +GKT + Y+L+ G + + P G + E
Sbjct: 306 TFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVE 458
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + + + DV G ++RP Y G++FVVD+ D A + L+ +L
Sbjct: 459 TVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLN 632
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + + LL+ NK D
Sbjct: 633 EDEL--RDAVLLVFANKQD 683
>TC76649 homologue to GP|20161472|dbj|BAB90396. ADP-ribosylation factor
{Oryza sativa (japonica cultivar-group)}, partial (48%)
Length = 1133
Score = 43.1 bits (100), Expect = 1e-04
Identities = 38/139 (27%), Positives = 64/139 (45%)
Frame = +1
Query: 41 TLLLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSE 100
T L RLF + K +++ GL +GKT + Y+L+ G + + P G + E
Sbjct: 130 TFTKLFSRLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVE 282
Query: 101 TTKKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILT 160
T + + + DV G ++RP Y G++FVVD+ D A + L+ +L
Sbjct: 283 TVEYKNIS-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLN 456
Query: 161 KGSVVKKKIPLLILCNKTD 179
+ + + LL+ NK D
Sbjct: 457 EDEL--RDAVLLVFANKQD 507
>TC76650 GP|965483|dbj|BAA08259.1| DcARF1 {Daucus carota}, complete
Length = 839
Score = 42.4 bits (98), Expect = 2e-04
Identities = 36/132 (27%), Positives = 62/132 (46%)
Frame = +2
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 119 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 271
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 272 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 439
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 440 DAVLLVFANKQD 475
>TC76380 homologue to PIR|T52341|T52341 ADP-ribosylation factor [imported] -
rice, complete
Length = 990
Score = 42.4 bits (98), Expect = 2e-04
Identities = 36/132 (27%), Positives = 62/132 (46%)
Frame = +1
Query: 48 RLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKV 107
RLF + K +++ GL +GKT + Y+L+ G + + P G + ET + +
Sbjct: 112 RLFAK-KEMRILMVGLDAAGKTTILYKLK-----LGEIVTTIPTIG---FNVETVEYKNI 264
Query: 108 KPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKK 167
+ DV G ++RP Y G++FVVD+ D A + L+ +L + + +
Sbjct: 265 S-FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSND-RDRVVEARDELHRMLNEDEL--R 432
Query: 168 KIPLLILCNKTD 179
LL+ NK D
Sbjct: 433 DAVLLVFANKQD 468
>TC78399 homologue to SP|O04834|SARA_ARATH GTP-binding protein SAR1A.
[Mouse-ear cress] {Arabidopsis thaliana}, complete
Length = 1164
Score = 42.4 bits (98), Expect = 2e-04
Identities = 44/153 (28%), Positives = 68/153 (43%), Gaps = 2/153 (1%)
Frame = +2
Query: 43 LLLSLRLFKRAKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETT 102
+L +L L++ K ++ GL +GKT L + L+D Q T +E I
Sbjct: 101 ILATLGLWQ--KEAKILFLGLDNAGKTTLLHMLKDERLVQHQPTQHPTSEELSI------ 256
Query: 103 KKGKVKPVHIVDVPGHSRLRPKLDEYLPQAAGVVFVVDAVD--FLPNCRAASEYLYDILT 160
G++K D+ GH R +Y Q VV++VDA D P + + L +
Sbjct: 257 --GRIK-FKAFDLGGHQIARRVWRDYYAQVDAVVYLVDAFDKERFPESKKELDALLADES 427
Query: 161 KGSVVKKKIPLLILCNKTDKVTAHTKEFIRRQL 193
G+V P LIL NK D A +++ +R L
Sbjct: 428 LGNV-----PFLILGNKIDITYAASEDELRYHL 511
>TC84764 similar to PIR|S71584|S71584 GTP-binding protein BG1 - Arabidopsis
thaliana, partial (79%)
Length = 705
Score = 39.7 bits (91), Expect = 0.001
Identities = 44/149 (29%), Positives = 70/149 (46%), Gaps = 7/149 (4%)
Frame = +1
Query: 58 VVLTGLTGSGKTVLFYQLRDGSTHQGTVTS--MEPNEGTFILHSETTKKGKVKPVHIVDV 115
V++ G+ +GKT L +++ T+ + + P G I E + V D+
Sbjct: 274 VLILGIDKAGKTTLLEKIKSVYTNVEGLPPDRIVPTVGLNIGRIEVANRKLV----FWDL 441
Query: 116 PGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVK----KKIPL 171
G LR ++Y +A VVFV+DA +C + E L K V++ K PL
Sbjct: 442 GGQLGLRSIWEKYYEEAHAVVFVIDA-----SCPSRFEDAKSALEK--VLRHEDLKGAPL 600
Query: 172 LILCNKTDKVTAHTKEFIRRQLE-KEIDK 199
LIL NK D A + E + R L+ K++D+
Sbjct: 601 LILANKQDLPEAVSSEELARYLDLKKLDE 687
>TC86809 homologue to PIR|S35701|S35701 translation elongation factor EF-G
chloroplast - soybean, partial (91%)
Length = 2674
Score = 36.6 bits (83), Expect = 0.010
Identities = 25/88 (28%), Positives = 40/88 (45%), Gaps = 7/88 (7%)
Frame = +2
Query: 62 GLTGSGKTVLFYQLRD---GSTHQGTVT----SMEPNEGTFILHSETTKKGKVKPVHIVD 114
G T + + +LFY R+ G H+GT T E G I + TT ++I+D
Sbjct: 353 GKTTTTERILFYTGRNYKIGEVHEGTATMDWMEQEQERGITITSAATTTFWDNHRINIID 532
Query: 115 VPGHSRLRPKLDEYLPQAAGVVFVVDAV 142
PGH +++ L G + + D+V
Sbjct: 533 TPGHVDFTLEVERALRVLDGAICLFDSV 616
>AW559547 weakly similar to GP|12654451|gb| ADP-ribosylation factor-like 7
{Homo sapiens}, partial (22%)
Length = 394
Score = 36.2 bits (82), Expect = 0.013
Identities = 36/122 (29%), Positives = 54/122 (43%)
Frame = +2
Query: 58 VVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHIVDVPG 117
VVL GL SGKT L + G + T L+ + KKG V+ + D+ G
Sbjct: 65 VVLVGLENSGKTTLLNVMAMGHPVETC--------PTIGLNVKLVKKGGVQ-MKCWDIGG 217
Query: 118 HSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTKGSVVKKKIPLLILCNK 177
++ R + Y +++VVDA F A + L+ +L + PLL+L NK
Sbjct: 218 QAQYRSEWGRYTRGCDVIIYVVDANAF-DQISLARKELHRLLEDRELA--TTPLLVLANK 388
Query: 178 TD 179
D
Sbjct: 389 ID 394
>TC90406 similar to GP|11994726|dbj|BAB03042. contains similarity to
ADP-ribosylation factor~gene_id:F5N5.14 {Arabidopsis
thaliana}, partial (98%)
Length = 912
Score = 35.8 bits (81), Expect = 0.017
Identities = 37/135 (27%), Positives = 56/135 (41%), Gaps = 2/135 (1%)
Frame = +2
Query: 53 AKTNTVVLTGLTGSGKTVLFYQLRDGSTHQGTVTSMEPNEGTFILHSETTKKGKVKPVHI 112
AK +V+ GL +GKT Y+L H G V + P G+ + E ++ +
Sbjct: 242 AKEYKIVVVGLDNAGKTTTLYKL-----HLGEVVTTNPTVGSNV---EELVYKNIR-FEV 394
Query: 113 VDVPGHSRLRPKLDEYLPQAAGVVFVVDAVDFLPNCRAASEYLYDILTK--GSVVKKKIP 170
D+ G RLR Y V+ V+D+ D RA + D L + G +
Sbjct: 395 WDLGGQERLRTSWATYYRGTHAVIAVIDSSD-----RARISLMKDELFRLLGHEDLQHSV 559
Query: 171 LLILCNKTDKVTAHT 185
+L+ NK D A T
Sbjct: 560 ILVFANKQDIKDAMT 604
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.318 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,583,523
Number of Sequences: 36976
Number of extensions: 94603
Number of successful extensions: 489
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 483
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 488
length of query: 259
length of database: 9,014,727
effective HSP length: 94
effective length of query: 165
effective length of database: 5,538,983
effective search space: 913932195
effective search space used: 913932195
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0008.13


