

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0008.11
(265 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
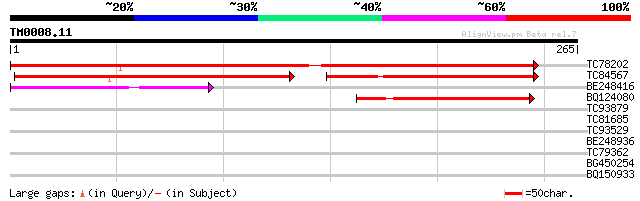
Score E
Sequences producing significant alignments: (bits) Value
TC78202 weakly similar to GP|16974602|gb|AAL31204.1 AT3g27760/MG... 349 5e-97
TC84567 similar to PIR|A86240|A86240 protein F20B24.10 [imported... 189 3e-74
BE248416 weakly similar to GP|15528715|dbj hypothetical protein~... 85 3e-17
BQ124080 weakly similar to GP|20197140|gb| predicted protein {Ar... 77 5e-15
TC93879 similar to GP|3877368|emb|CAA92514.1 cDNA EST EMBL:T0125... 29 1.6
TC81685 27 6.2
TC93529 similar to GP|15391731|emb|CAA93316. nitrite transporter... 27 6.2
BE248936 27 8.1
TC79362 WD-repeat cell cycle regulatory protein 27 8.1
BG450254 similar to PIR|T48059|T4 ABC transporter-like protein -... 27 8.1
BQ150933 27 8.1
>TC78202 weakly similar to GP|16974602|gb|AAL31204.1 AT3g27760/MGF10_16
{Arabidopsis thaliana}, partial (33%)
Length = 1541
Score = 349 bits (896), Expect = 5e-97
Identities = 170/248 (68%), Positives = 197/248 (78%), Gaps = 1/248 (0%)
Frame = +1
Query: 1 METDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGER-QQQRAGLLY 59
M DT LSYWLNWRF ICA++++ TMGLGSFLIWKYEEFNKSR+ER E +++ GLLY
Sbjct: 178 MTPDTNTLSYWLNWRFFICAIFLLLTMGLGSFLIWKYEEFNKSRNERVEEGRRETVGLLY 357
Query: 60 EDELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFG 119
EDE WNTC+K IHP WLL+YRI+SF VLLGLL N+V DG GI YFYTQWTFTLVTIYF
Sbjct: 358 EDEAWNTCVKGIHPNWLLSYRIISFFVLLGLLTANVVVDGGGIFYFYTQWTFTLVTIYFA 537
Query: 120 LASFFSIYGCFFKHDKFEGNTVGSACLNTERGTYVAPTIDGDTDIPHMYKSTDAYQEPHT 179
LAS FS Y F H++FEGNT L+ ERGTYVAPT+DG +DIP + KS+ + +E
Sbjct: 538 LASCFSFYRSCFNHNEFEGNT-----LDRERGTYVAPTLDGISDIPVLSKSSYSNRESLN 702
Query: 180 QNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLL 239
+NTAG WGYI QI+FQTCAGA +LTDLVFWLV+YPF+ KDFRL +F V MHSVNAV LL
Sbjct: 703 RNTAGVWGYIIQILFQTCAGAAMLTDLVFWLVIYPFMTSKDFRLDIFTVGMHSVNAVLLL 882
Query: 240 GETSLNGM 247
GETSLN M
Sbjct: 883 GETSLNCM 906
Score = 31.6 bits (70), Expect = 0.33
Identities = 25/71 (35%), Positives = 37/71 (51%)
Frame = +1
Query: 192 IMFQTCAGAVVLTDLVFWLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGETSLNGMVKYL 251
++FQ AVV ++W YPFL L+ + + + + G +L +K+L
Sbjct: 958 VIFQWIVHAVVS---LWWP--YPFLDLSSPYAPLWYLAV-GIMHIPCYGFFALIVKLKHL 1119
Query: 252 WLSRLFPGSCQ 262
WLSRLF GSCQ
Sbjct: 1120 WLSRLFSGSCQ 1152
>TC84567 similar to PIR|A86240|A86240 protein F20B24.10 [imported] -
Arabidopsis thaliana, partial (16%)
Length = 1305
Score = 189 bits (479), Expect(2) = 3e-74
Identities = 88/132 (66%), Positives = 100/132 (75%), Gaps = 1/132 (0%)
Frame = +1
Query: 3 TDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRD-ERGERQQQRAGLLYED 61
TDTTALSYWLNWRF CALWI +M L S+LI+KYE FNK R ER E Q+ GLLYED
Sbjct: 175 TDTTALSYWLNWRFFFCALWIFISMILASYLIFKYEGFNKERSSERDENHQEEDGLLYED 354
Query: 62 ELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLA 121
E WNTC+K I P+WLL YRI+SF++LL L+I N+ G IL +YTQ TFTLVTIYFGL
Sbjct: 355 EAWNTCVKGIDPIWLLVYRIISFVILLALIIANVAVSGASILAYYTQLTFTLVTIYFGLG 534
Query: 122 SFFSIYGCFFKH 133
S FSIYGC KH
Sbjct: 535 SSFSIYGCLLKH 570
Score = 107 bits (267), Expect(2) = 3e-74
Identities = 53/99 (53%), Positives = 69/99 (69%)
Frame = +3
Query: 149 ERGTYVAPTIDGDTDIPHMYKSTDAYQEPHTQNTAGAWGYIFQIMFQTCAGAVVLTDLVF 208
++ Y+AP+++ DIP + K + QE HT+ AG WGYI QI++QTCAGAV LTD VF
Sbjct: 615 QKALYMAPSLERVLDIPLLPKCPN--QEFHTREIAGVWGYISQIIYQTCAGAVFLTDFVF 788
Query: 209 WLVLYPFLAPKDFRLGLFAVCMHSVNAVFLLGETSLNGM 247
W VLYP A + +L CMH++NAVFLLG+TSLN M
Sbjct: 789 WFVLYPVRAFNNDKLDFLNFCMHTINAVFLLGDTSLNCM 905
>BE248416 weakly similar to GP|15528715|dbj hypothetical protein~similar to
Arabidopsis thaliana chromosome 5 MJH22.1, partial (5%)
Length = 335
Score = 84.7 bits (208), Expect = 3e-17
Identities = 42/95 (44%), Positives = 53/95 (55%)
Frame = +3
Query: 1 METDTTALSYWLNWRFSICALWIIFTMGLGSFLIWKYEEFNKSRDERGERQQQRAGLLYE 60
M+ D+T LSYWLNWR IC + ++ +M L IWK E + GE Q G
Sbjct: 63 MQLDSTTLSYWLNWRVYICVVSVLLSMILACLTIWKRESSRNLTFDNGENQSTLCG---- 230
Query: 61 DELWNTCLKRIHPVWLLAYRIVSFIVLLGLLIGNM 95
DE W CLK IHPV LL +R++SF LL LI +
Sbjct: 231 DEAWKPCLKDIHPVCLLVFRVISFSSLLASLIAKI 335
>BQ124080 weakly similar to GP|20197140|gb| predicted protein {Arabidopsis
thaliana}, partial (22%)
Length = 660
Score = 77.4 bits (189), Expect = 5e-15
Identities = 34/83 (40%), Positives = 53/83 (62%)
Frame = +1
Query: 163 DIPHMYKSTDAYQEPHTQNTAGAWGYIFQIMFQTCAGAVVLTDLVFWLVLYPFLAPKDFR 222
++P +++ Q+ A W YIFQI+FQ AGAV+LTD ++WL+++PFL +D+
Sbjct: 16 NVPFLHQDATNIQK---NQIAPVWSYIFQILFQISAGAVMLTDCIYWLIIFPFLTLRDYD 186
Query: 223 LGLFAVCMHSVNAVFLLGETSLN 245
L V MH++N V L G+ +LN
Sbjct: 187 LNFMTVNMHTLNLVLLFGDAALN 255
>TC93879 similar to GP|3877368|emb|CAA92514.1 cDNA EST EMBL:T01259 comes
from this gene~cDNA EST yk67d3.3 comes from this
gene~cDNA EST yk67d3.5, partial (68%)
Length = 426
Score = 29.3 bits (64), Expect = 1.6
Identities = 15/49 (30%), Positives = 24/49 (48%)
Frame = -3
Query: 69 KRIHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIY 117
+ +H VWL ++ F++LL + LY+Y W F +V IY
Sbjct: 184 QEVHLVWLFLLILLVFLLLLAVFS*TQRHFHPNHLYYYLFWFF*VVLIY 38
>TC81685
Length = 1706
Score = 27.3 bits (59), Expect = 6.2
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 1/39 (2%)
Frame = +1
Query: 158 IDGDTDIPHMY-KSTDAYQEPHTQNTAGAWGYIFQIMFQ 195
+DGD PHM+ S+D Q+ +N + W + Q+ Q
Sbjct: 1120 MDGDDFEPHMFHHSSDIQQDYRMENMSKKWWCVMQVFQQ 1236
>TC93529 similar to GP|15391731|emb|CAA93316. nitrite transporter {Cucumis
sativus}, partial (10%)
Length = 423
Score = 27.3 bits (59), Expect = 6.2
Identities = 12/41 (29%), Positives = 20/41 (48%)
Frame = +1
Query: 87 LLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASFFSIY 127
++G LIG ++ + Y+ L IYFG+ +F Y
Sbjct: 55 IIGYLIGTLIRGKLEYYYYLVSGVQVLNLIYFGICFWFYTY 177
>BE248936
Length = 444
Score = 26.9 bits (58), Expect = 8.1
Identities = 19/62 (30%), Positives = 28/62 (44%)
Frame = -3
Query: 71 IHPVWLLAYRIVSFIVLLGLLIGNMVADGVGILYFYTQWTFTLVTIYFGLASFFSIYGCF 130
IH +LL+YR V F + L ++ + +Y Q T YF FF++ CF
Sbjct: 364 IHKRFLLSYRTVKFAKRIHL---SLFLIFI*SIYLLNQLTSLNNPNYFNYLLFFTLTTCF 194
Query: 131 FK 132
K
Sbjct: 193 HK 188
>TC79362 WD-repeat cell cycle regulatory protein
Length = 1800
Score = 26.9 bits (58), Expect = 8.1
Identities = 11/31 (35%), Positives = 18/31 (57%)
Frame = +2
Query: 145 CLNTERGTYVAPTIDGDTDIPHMYKSTDAYQ 175
C T + T++ P D +IPH K+ D+Y+
Sbjct: 1331 CQ*TSKHTWLLPEPDNCLEIPHYVKAGDSYR 1423
>BG450254 similar to PIR|T48059|T4 ABC transporter-like protein - Arabidopsis
thaliana, partial (10%)
Length = 616
Score = 26.9 bits (58), Expect = 8.1
Identities = 10/45 (22%), Positives = 25/45 (55%), Gaps = 3/45 (6%)
Frame = +1
Query: 68 LKRIHPVWLLAYRIVSFIVLLGLLIGNMVAD---GVGILYFYTQW 109
+K+ HP+W++ ++ + + L+ +G V G I++ +T +
Sbjct: 301 MKQFHPIWMMPLQVAAALALMYSYVGVSVVAAILGTAIVFCFTAY 435
>BQ150933
Length = 677
Score = 26.9 bits (58), Expect = 8.1
Identities = 13/37 (35%), Positives = 17/37 (45%), Gaps = 7/37 (18%)
Frame = +1
Query: 105 FYTQWTFTLVTIY-------FGLASFFSIYGCFFKHD 134
F+ W F + Y F A+FFS + CF HD
Sbjct: 82 FFYFWPFLIALSYSPYFT*AFLSAAFFSFFSCFLLHD 192
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.328 0.143 0.473
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,666,474
Number of Sequences: 36976
Number of extensions: 163260
Number of successful extensions: 1205
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 1191
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1200
length of query: 265
length of database: 9,014,727
effective HSP length: 94
effective length of query: 171
effective length of database: 5,538,983
effective search space: 947166093
effective search space used: 947166093
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0008.11


