

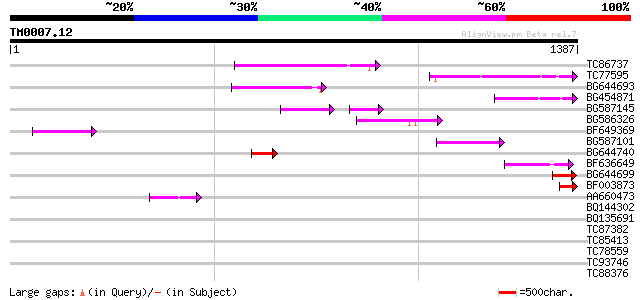
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0007.12
(1387 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alterna... 240 3e-63
TC77595 weakly similar to PIR|T18350|T18350 probable pol polypro... 164 2e-40
BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin... 161 2e-39
BG454871 weakly similar to GP|10140673|g putative gag-pol polypr... 117 4e-26
BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - ... 97 7e-26
BG586326 similar to PIR|G84493|G8 probable retroelement pol poly... 97 3e-20
BF649369 87 4e-17
BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana... 82 1e-15
BG644740 similar to PIR|A84460|A84 probable retroelement pol pol... 66 1e-10
BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativ... 58 2e-08
BG644699 similar to PIR|T07863|T078 probable polyprotein - pinea... 53 9e-07
BF003873 similar to GP|14715222|em putative polyprotein {Cicer a... 49 1e-05
AA660473 44 4e-04
BQ144302 similar to GP|6815109|dbj| hypothetical protein {Oryza ... 39 0.018
BQ135691 similar to PIR|E96636|E966 hypothetical protein T7P1.21... 37 0.069
TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulch... 34 0.34
TC85413 similar to PIR|S23737|S23737 proline-rich protein precur... 34 0.45
TC78559 weakly similar to GP|5733686|gb|AAD49719.1| maturation p... 33 0.59
TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical... 33 0.59
TC88376 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidop... 32 1.3
>TC86737 weakly similar to GP|6683624|dbj|BAA89272.1 Pol {Alternaria
alternata}, partial (21%)
Length = 1540
Score = 240 bits (612), Expect = 3e-63
Identities = 146/372 (39%), Positives = 209/372 (55%), Gaps = 13/372 (3%)
Frame = +1
Query: 549 DHHINLLP----NTPPVNVRPYRYPHSQKEAMAT--ILTDMLQEGIVVPSTSPYSSPVLL 602
DH I L+P N PP+ P Y S++E + L D+L +G + S S +PVL
Sbjct: 415 DHAIPLIPDKDGNDPPLPWGPL-YGMSRQELLVLKKTLEDLLDKGFIKASGSAAGAPVLF 591
Query: 603 IKKKDGTWRFCVDFRSLNSITIKDRFPIPTIDELLDELGGASHFSKLDLRSGFHQIRLAT 662
++K G RFCVD+R+LN+IT KDR+P+P I E L + GA F+KLD+ + FH++R+
Sbjct: 592 VRKPGGGIRFCVDYRALNAITKKDRYPLPLISETLRRVAGARWFTKLDVVAAFHKMRIKD 771
Query: 663 EDTHKTAFRTVDGHYEFLVMPFGLTNAPSTFQAAMNDLLRPFLRRFVLVFFDDILVYSP- 721
ED KTAFRT G +E++V PFGLT AP+TFQ +N L FL FV + DD+L+Y+
Sbjct: 772 EDQEKTAFRTRYGLFEWIVCPFGLTGAPATFQRYINKTLHEFLDDFVTAYIDDVLIYTTG 951
Query: 722 SLSAHMTHLKEVLEVLLSHKFYAKLSKCIFGVTSVSYLGHIISA-NGVGPDPSKVQAIVD 780
S H ++ VL L KC F VT+V Y+G I++A GV DP K+ AI D
Sbjct: 952 SKKDHEAQVRRVLRRLADAGLSLDPKKCEFSVTTVKYVGFILTAGKGVSCDPLKLAAIRD 1131
Query: 781 WPVPRNLTALRGFLGLTGFYRRFIKNYAAHASHLTDLLKQDQKDKHTLLPWSSAAADSFQ 840
W P ++ R FLG +Y+ FI Y+ LT L ++D + W + +F
Sbjct: 1132WLPPGSVKGARSFLGFCNYYKDFIPGYSEITEPLTRLTRKDFPFR-----WGAEQEAAFT 1296
Query: 841 HLKDLIISAPVLVLPDFSATFDIETDASGTAVGAVLSQK-----GHPISFFSKKLTLQMQ 895
LK L PVL + D A +ETD SG A+G VL+Q+ HP++F S++L+
Sbjct: 1297KLKRLFAEEPVLRMFDPEAVTTVETDCSGFALGGVLTQEDGTGAAHPVAFHSQRLSPAEY 1476
Query: 896 HQSTYVREMYAV 907
+ + +E+ AV
Sbjct: 1477NYPIHDKELLAV 1512
>TC77595 weakly similar to PIR|T18350|T18350 probable pol polyprotein - rice
blast fungus gypsy retroelement (fragment), partial (14%)
Length = 1708
Score = 164 bits (415), Expect = 2e-40
Identities = 124/378 (32%), Positives = 181/378 (47%), Gaps = 17/378 (4%)
Frame = +2
Query: 1027 LYFKDGLFVPEHD-------QWRTSILSEYHASPAAGHSGLKPTLARLMASFNWPGIQTE 1079
L F+ ++VP D + RT ++ E H S AAGH G TL + F WPG
Sbjct: 104 LTFRGRIWVPGSDDEESPLNELRTKLVQESHDSTAAGHPGRNGTLEIVSRKFFWPGQSQT 283
Query: 1080 TKTFIKQCLPCQYNKYVPAKKSGLLQPLPTPAQIWEDISMDFITGLPPSHGH--TVAWVI 1137
+ F++ C C K G L+PLP P ++ D+SMDFIT LPP+ G WVI
Sbjct: 284 VRRFVRNCDVCGGIHIWRQAKRGFLKPLPVPNRLHSDLSMDFITSLPPTRGRGSQYLWVI 463
Query: 1138 VDRLSKYAHFVALPANFTATSLANRFSSEICRLHGIPRSIVSDRDKIFLSHFWRDLFRVY 1197
VDRLSK + A + A RF S R HG+P+SIVSDR ++ FWR+ R+
Sbjct: 464 VDRLSKSVTLEEMDT-MEAEACAQRFLSCHYRFHGMPQSIVSDRGSNWVGRFWREFCRLT 640
Query: 1198 GTKLRFSTAYHPETDGQTEVVNRGLETYLRCFAGEQPRSWYKFLHLAELWYNTSFHSAAG 1257
G ST+YHP+TDG TE N+ ++ LR + +W L +L +S+ G
Sbjct: 641 GVTQLLSTSYHPQTDGGTERWNQEIQAVLRAYVCWSQDNWGDLLPTVQLALRNRHNSSIG 820
Query: 1258 MTPFQAVYG---RPPPSLL----AYVPGSSAIQSLDESLQQRT*ILESLKANLQRAQHRM 1310
TPF +G P P++ G +A Q L + ++ T ++ A + AQ R
Sbjct: 821 ATPFFVEHGYHVDPIPTVEDTGGVVSEGEAAAQLLVKRMKDVTGFIQ---AEIVAAQQRS 991
Query: 1311 KIQKDKSRREV-TFEENAWVLLRLQPYRQRSLAHRLSNKLAKRFYGPFRVKRRIGSVAYE 1369
+ +K R ++ V L + Y+ + R S KL + + V R + E
Sbjct: 992 EASANKRRCPADRYQVGDKVWLNVSNYK----SPRPSKKL-DWLHHKYEVTRFVTPHVVE 1156
Query: 1370 LDLPPTSKLHNVFHVSLL 1387
L++P T ++ FHV LL
Sbjct: 1157 LNVPGT--VYPKFHVDLL 1204
>BG644693 weakly similar to GP|18767374|g Putative 22 kDa kafirin cluster;
Ty3-Gypsy type {Oryza sativa}, partial (15%)
Length = 716
Score = 161 bits (407), Expect = 2e-39
Identities = 94/243 (38%), Positives = 132/243 (53%), Gaps = 10/243 (4%)
Frame = +2
Query: 542 LPPTRFHDHHINLLPNTPPVNVRPYRYPHSQKEAMATILTDMLQEGIVVPSTSPYSSPVL 601
+PP D I+LLPN P+ + YR + + + L D+L++G + PS P VL
Sbjct: 17 VPPEWKIDFGIDLLPNMNPI*IPSYRINPLKLKVLKLQLKDLLEKGFIQPSIYP*GVVVL 196
Query: 602 LIKKKDGTWRFCVDFRSLNSITIKDRFPIPTIDELLDELGGASHFSKLDLRSGFHQIRLA 661
+KKKDG R +D+ LN++ IK ++P+P IDEL D L G+ F K+DLR G HQ R+
Sbjct: 197 FLKKKDGFLRMSIDYPQLNNVNIKIKYPLPLIDELFDNLQGSKWFFKIDLRLG*HQHRVI 376
Query: 662 TEDTHKTAFRTVDGHYEFLVMPFGLTNAPSTFQAAMNDLLRPFLRRFVLVFFDDILVYSP 721
ED KTAFR GHYE LVM FG TN P F MN + + +L V+VF +DIL+YS
Sbjct: 377 GEDVPKTAFRIRYGHYEILVMSFG*TNPPMAFMELMNRVFQDYLDSLVIVFSNDILIYSK 556
Query: 722 SLSAHMTHLKEVLEVLLSHKFYAKLSKCIFGVTSVSYL----------GHIISANGVGPD 771
+ + H HL+ L+VL G+ +SY+ H+IS G+ D
Sbjct: 557 NENEHENHLRLALKVLKD-----------IGLCQISYV*ILVEVGFFSLHVISGEGLKVD 703
Query: 772 PSK 774
+
Sbjct: 704 SKR 712
>BG454871 weakly similar to GP|10140673|g putative gag-pol polyprotein {Oryza
sativa (japonica cultivar-group)}, partial (7%)
Length = 674
Score = 117 bits (292), Expect = 4e-26
Identities = 79/203 (38%), Positives = 104/203 (50%), Gaps = 2/203 (0%)
Frame = +2
Query: 1187 SHFWRDLFRVYGTKLRFSTAYHPETDGQTEVVNRGLETYLRCFAGEQPRSWYKFLHLAEL 1246
S+FW+ LF+++GT L S+AYHP +DGQ+E +N+G E YLRC P W K AE
Sbjct: 32 SNFWKQLFKLHGTILTMSSAYHP*SDGQSEALNKGXEMYLRCLMFTDPLKWSKAFPWAEY 211
Query: 1247 WYNTSFHSAAGMTPFQAVYGRPPPSLLAYVPGSSAIQSLDESLQQRT*ILESLKANLQRA 1306
WYNTS++ +A MTPF+A+YGR L+ S L L QR E L + LQ
Sbjct: 212 WYNTSYNISAAMTPFKALYGRDLSMLIRSKGSSKDTADLQSQLAQR----EELLSQLQSI 379
Query: 1307 QHRM-KIQKDKSRREVTFEENAWVLLRL-QPYRQRSLAHRLSNKLAKRFYGPFRVKRRIG 1364
R+ K+ K R ++WV L SL N K P V
Sbjct: 380 STRLNKL*SIKLIRNAAILSSSWVSTSL*NCNLINSLR*HCGN--IKSSVHPTLV-HY*Q 550
Query: 1365 SVAYELDLPPTSKLHNVFHVSLL 1387
S AY+L LP T+K+ +FHVS L
Sbjct: 551 SAAYKLSLPSTAKVPPIFHVSQL 619
>BG587145 similar to PIR|H86337|H8 protein F5M15.26 [imported] - Arabidopsis
thaliana, partial (13%)
Length = 763
Score = 97.1 bits (240), Expect(2) = 7e-26
Identities = 52/133 (39%), Positives = 77/133 (57%)
Frame = +2
Query: 663 EDTHKTAFRTVDGHYEFLVMPFGLTNAPSTFQAAMNDLLRPFLRRFVLVFFDDILVYSPS 722
+D KTAF T G Y + VMPFGL NA ST+Q +N + L + V+ DD+LV S
Sbjct: 11 DDLEKTAFITDRGTYCYKVMPFGLKNAGSTYQRLVNRMFADKLGNTMEVYIDDMLVKSLR 190
Query: 723 LSAHMTHLKEVLEVLLSHKFYAKLSKCIFGVTSVSYLGHIISANGVGPDPSKVQAIVDWP 782
+ H+ HLKE + L + +KC FGVTS +LG+I++ G+ +P ++ AI+D P
Sbjct: 191 ATDHLNHLKE*FKTLDEYIMKLNPAKCTFGVTSGEFLGYIVTQQGIEVNPKQITAILDLP 370
Query: 783 VPRNLTALRGFLG 795
P+N ++ G
Sbjct: 371 SPKNSREVQRLTG 409
Score = 40.0 bits (92), Expect(2) = 7e-26
Identities = 24/87 (27%), Positives = 41/87 (46%), Gaps = 4/87 (4%)
Frame = +3
Query: 831 WSSAAADSFQHLKDLIISAPVLVLPDFSATFDIETDASGTAVGAVLSQKGH----PISFF 886
W ++F+ LK + + PVL P+ T + S TAV +VL ++ PI +
Sbjct: 501 WDEKCEEAFEQLKQYLTTPPVLSKPEAGDTLSLYIAISSTAVSSVLIREDRGEQKPIFYT 680
Query: 887 SKKLTLQMQHQSTYVREMYAVTEAVKK 913
SK++T T + +AV + +K
Sbjct: 681 SKRMTDPETRYPTLEKMAFAVITSARK 761
>BG586326 similar to PIR|G84493|G8 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (13%)
Length = 736
Score = 97.4 bits (241), Expect = 3e-20
Identities = 70/236 (29%), Positives = 109/236 (45%), Gaps = 24/236 (10%)
Frame = +2
Query: 848 SAPVLVLPDFSATFDIETDASGTAVGAVLSQKGHPISFFSKKLTLQMQHQSTYVREMYAV 907
SAP+LVLP+ T+ + TDAS T +G VL+Q I++ S++L + T+ EM AV
Sbjct: 11 SAPILVLPEL-ITYVVYTDASITGLGCVLTQHEKVIAYASRQLRKHEGNYPTHDLEMAAV 187
Query: 908 TEAVKKWRQYLIGHKFRIYTDQQSLKHLMTQTFQTPDQIKWATKLLGFDYEIFYKPGSEN 967
A+K WR YL G K +I+TD +SLK++ TQ Q +W + +D +I Y PG N
Sbjct: 188 VFALKIWRSYLYGAKVQIHTDHKSLKYIFTQPELNLRQRRWMEFVADYDLDITYYPGKAN 367
Query: 968 RVADALSR-----------------CHSSELPLLAAISSPV-------PEIITQLKQYYK 1003
VADALSR + L +L + + ++ T+++
Sbjct: 368 LVADALSRRRVDVSAEREADDLDGMVRALRLNVLTKATESLGLEAVNQADLFTRIRLAQG 547
Query: 1004 TPEGIQLIVDKSTLPHFRVHHEVLYFKDGLFVPEHDQWRTSILSEYHASPAAGHSG 1059
E +Q + + + + VP + I+SE H S + H G
Sbjct: 548 QDENLQKVAQNDRTEYQTAKDGTILVNGRISVPNDRSLKEEIMSEAHKSRFSVHPG 715
>BF649369
Length = 631
Score = 87.0 bits (214), Expect = 4e-17
Identities = 50/157 (31%), Positives = 79/157 (49%)
Frame = +3
Query: 56 KLQLTSFDGSKPLEWLFHAEQFFQLHQTPPAQRLELVSFYMQGDALSWYRWMHNNYQLSF 115
K++L F+G P+ W+ AE +F + TP R++L M+G + W+ +
Sbjct: 123 KVKLPLFEGDDPVAWITRAEIYFDVQNTPDDMRVKLSRLSMEGPTIHWFNLLMETEDDLS 302
Query: 116 WEAFTRALAIRFGPSAYENHQQELFKLQQTSTVAEYQTRFEKVSNQVVGLSPEAVLDCFI 175
E +AL R+ EN +EL L+Q +V E+ FE +S+QV L E L F+
Sbjct: 303 REKLKKALIARYDGRRLENPFEELSTLRQIGSVEEFVEAFELLSSQVGRLPEEQYLGYFM 482
Query: 176 SGLVPEIKRELAILKPYSVSQAIGIAKQIEAKIKESR 212
SGL I+R + L P + Q + IAK +E + + R
Sbjct: 483 SGLKAHIRRRVRTLNPTTRMQMMRIAKDVEDEFERGR 593
>BG587101 similar to GP|6691191|gb F7F22.15 {Arabidopsis thaliana}, partial
(10%)
Length = 624
Score = 82.0 bits (201), Expect = 1e-15
Identities = 49/167 (29%), Positives = 86/167 (51%), Gaps = 1/167 (0%)
Frame = +2
Query: 1045 ILSEYHASPAAGHSGLKPTLARLM-ASFNWPGIQTETKTFIKQCLPCQYNKYVPAKKSGL 1103
IL H S AGH + T++++ A F WP + + +FI +C PCQ + + ++ +
Sbjct: 104 ILFHCHGSNYAGHFAVSKTVSKIQQAGFWWPTMFKDAHSFISKCDPCQRQGNI-S*RNEM 280
Query: 1104 LQPLPTPAQIWEDISMDFITGLPPSHGHTVAWVIVDRLSKYAHFVALPANFTATSLANRF 1163
Q ++++ +DF+ P S+ + V VD +SK+ +A P N AT + F
Sbjct: 281 PQNFILEVEVFDVWGIDFMGPFPSSYNNKYILVAVDYVSKWVEAIASPTN-DATVVVKMF 457
Query: 1164 SSEICRLHGIPRSIVSDRDKIFLSHFWRDLFRVYGTKLRFSTAYHPE 1210
S I G+PR ++SD F++ + L + G + + +TAYHP+
Sbjct: 458 KSVIFPRFGVPRVVISDGGSHFINKVFEKLLKKNGVRHKVATAYHPQ 598
>BG644740 similar to PIR|A84460|A84 probable retroelement pol polyprotein
[imported] - Arabidopsis thaliana, partial (4%)
Length = 754
Score = 65.9 bits (159), Expect = 1e-10
Identities = 27/64 (42%), Positives = 43/64 (67%)
Frame = -1
Query: 591 PSTSPYSSPVLLIKKKDGTWRFCVDFRSLNSITIKDRFPIPTIDELLDELGGASHFSKLD 650
PS SP + +L ++KKDG +R C+D+R N +T K+++P+P ID L D++ +F +D
Sbjct: 259 PSISP*GAALLFVRKKDGYFRMCIDYRQFNKVTTKNKYPLPRIDNLFDKIQEDCYF*NID 80
Query: 651 LRSG 654
LR G
Sbjct: 79 LRLG 68
>BF636649 similar to GP|21628724|emb OSJNBa0033H08.7 {Oryza sativa}, partial
(4%)
Length = 653
Score = 58.2 bits (139), Expect = 2e-08
Identities = 51/170 (30%), Positives = 74/170 (43%)
Frame = +1
Query: 1210 ETDGQTEVVNRGLETYLRCFAGEQPRSWYKFLHLAELWYNTSFHSAAGMTPFQAVYGRPP 1269
++D Q ++N LET+L F EQ FL AE YNT+FH AG TPF+ VY
Sbjct: 4 DSDEQAGLLNHTLETHLLYFTSEQQGV*NFFLTWAECLYNTNFHRTAGCTPFKVVY--VV 177
Query: 1270 PSLLAYVPGSSAIQSLDESLQQRT*ILESLKANLQRAQHRMKIQKDKSRREVTFEENAWV 1329
L +V I + + T* A R + R
Sbjct: 178 AHLQKFVVARDLIYRNEGLHKSST*TSFGRGTRAYEALSRPAYETC*HPRRP-------- 333
Query: 1330 LLRLQPYRQRSLAHRLSNKLAKRFYGPFRVKRRIGSVAYELDLPPTSKLH 1379
L + R R+ ++ K YGP++V ++IGSVA++L LP ++H
Sbjct: 334 -LSIVYTRDRTYEWQVLPKYVA*CYGPYQVIKQIGSVAFKL*LPEQHQIH 480
>BG644699 similar to PIR|T07863|T078 probable polyprotein - pineapple
retrotransposon dea1 (fragment), partial (5%)
Length = 231
Score = 52.8 bits (125), Expect = 9e-07
Identities = 29/59 (49%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Frame = +2
Query: 1329 VLLRLQPYRQRSLAHRLSNKLAKRFYGPFRVKRRIGSVAYELDLPP-TSKLHNVFHVSL 1386
VLL++ P + KL+ R+ GPF V +RIG VAYEL LPP S +H VFHVS+
Sbjct: 8 VLLKVLPTERGDCRFGKRGKLSLRYIGPFEVIKRIGEVAYELALPPGLSGVHPVFHVSM 184
>BF003873 similar to GP|14715222|em putative polyprotein {Cicer arietinum},
partial (82%)
Length = 559
Score = 48.9 bits (115), Expect = 1e-05
Identities = 23/43 (53%), Positives = 30/43 (69%), Gaps = 1/43 (2%)
Frame = +2
Query: 1346 SNKLAKRFYGPFRVKRRIGSVAYELDLPP-TSKLHNVFHVSLL 1387
S KL RF GP+++ R+G+VAY + LPP LH+VFHVS L
Sbjct: 23 SKKLTVRFIGPYQISERVGTVAYRVGLPPHLLNLHDVFHVSQL 151
>AA660473
Length = 655
Score = 43.9 bits (102), Expect = 4e-04
Identities = 39/130 (30%), Positives = 62/130 (47%), Gaps = 4/130 (3%)
Frame = -2
Query: 343 PRTLKFTAIVNGHPVVVLVDTGSSNNFVPPRTVSFLHLKVTPIPTFPVMVGNGAHIPCAG 402
P TL+ + G P+V+LVD G+++NF +VS + K I + VG + G
Sbjct: 549 PATLQLVGCLKGVPIVILVDIGANHNFDFASSVSDVGCKGRVILSQESEVG*WSENINVG 370
Query: 403 YIPDIEVTFAGNTFH---IPFYVMDLQGADFVLGLDWLKTLGK-VISDFSIPSMSFVVNG 458
T G FH + Y ++L D LG+ WL+ LG V+ D+ ++ F G
Sbjct: 369 *AY*SRGTVRG--FHC*GVDAYELELGEFDMFLGVAWLEKLGN*VVFDWDERTICFEWKG 196
Query: 459 KTCTLEGEPL 468
+ L+G+ L
Sbjct: 195 EVVKLQGQIL 166
>BQ144302 similar to GP|6815109|dbj| hypothetical protein {Oryza sativa
(japonica cultivar-group)}, partial (3%)
Length = 1047
Score = 38.5 bits (88), Expect = 0.018
Identities = 29/100 (29%), Positives = 40/100 (40%), Gaps = 2/100 (2%)
Frame = +1
Query: 469 PPPSHASFNHFQRLIHTDAIAECHTITFLPSPPSTPSPQFLTLENLSTPPPDFDPALWEL 528
PPP F LI + C + F+ SP P P L+ + PPP F P
Sbjct: 358 PPPPFPFFLFVSPLIFFHTVCPCFSFNFIISP-LLPPPTLLSHISPFPPPPFFSPPFHHF 534
Query: 529 LQS--YAPVFSTPHGLPPTRFHDHHINLLPNTPPVNVRPY 566
S + P FS L P F ++L + P V++ Y
Sbjct: 535 SSSPHFLPFFSFFPYLSPFHFPHPFLHLFSSYPFVSIFSY 654
>BQ135691 similar to PIR|E96636|E966 hypothetical protein T7P1.21 [imported]
- Arabidopsis thaliana, partial (4%)
Length = 1215
Score = 36.6 bits (83), Expect = 0.069
Identities = 29/78 (37%), Positives = 39/78 (49%), Gaps = 7/78 (8%)
Frame = -2
Query: 498 PSPPSTPSPQFLTLENLSTPPPD-FDP----ALWELLQSYAPVFST--PHGLPPTRFHDH 550
P PP+T SP +T TPPP +DP A+ L + P+ ST P+ P R + H
Sbjct: 878 PPPPTTISPHTIT----RTPPPTAYDPFTARAIPALPSPH*PISSTLTPYTRAP*RTYTH 711
Query: 551 HINLLPNTPPVNVRPYRY 568
+ L P+ PP P RY
Sbjct: 710 PVPLPPSAPP---SPTRY 666
>TC87382 similar to EGAD|146423|156195 vitellogenin {Anolis pulchellus},
partial (7%)
Length = 2304
Score = 34.3 bits (77), Expect = 0.34
Identities = 19/88 (21%), Positives = 42/88 (47%), Gaps = 10/88 (11%)
Frame = +2
Query: 55 LKLQLTSFDGSKPLE----WLFHAEQFFQLHQTPPAQRLELVSFYMQGDALSWY------ 104
+K+ + F+G+ L+ WL E+ F+ + P Q++++V+ ++ AL W+
Sbjct: 614 IKVDIPDFEGNLQLDDFLDWLQTIERVFEYKEVPEEQKVKIVAAKLKKHALIWWENLKRR 793
Query: 105 RWMHNNYQLSFWEAFTRALAIRFGPSAY 132
R ++ W+ + L ++ P Y
Sbjct: 794 RKREGKSKIKTWDKMRQKLTRKYLPPHY 877
>TC85413 similar to PIR|S23737|S23737 proline-rich protein precursor -
kidney bean, partial (56%)
Length = 1139
Score = 33.9 bits (76), Expect = 0.45
Identities = 28/97 (28%), Positives = 37/97 (37%), Gaps = 1/97 (1%)
Frame = +1
Query: 467 PLPPPSHASFNHFQRLIHTDAIAECHTITFLPSPPSTPSPQFLTLENLSTPPPDFDPALW 526
PL PP++A +H IH+ P+P TP+P +P P P
Sbjct: 160 PLHPPANAPHHHHHHHIHS------------PTPAPTPTP---------SPSPIHTPL-- 270
Query: 527 ELLQSYAPVFSTP-HGLPPTRFHDHHINLLPNTPPVN 562
+ P S P PPT H HH P PV+
Sbjct: 271 -----HPPYHSAPVPAKPPTHGHHHHHPHPPAPTPVH 366
>TC78559 weakly similar to GP|5733686|gb|AAD49719.1| maturation protein
pPM32 {Glycine max}, partial (55%)
Length = 1128
Score = 33.5 bits (75), Expect = 0.59
Identities = 31/98 (31%), Positives = 42/98 (42%), Gaps = 3/98 (3%)
Frame = -2
Query: 497 LPSPPSTPSPQFLT--LENLSTPPPDFDPALWELLQSYAPVFSTPHGLPPTRFHDHHINL 554
L P S+P PQ + L PPP P LL SY P + L P F HH +
Sbjct: 734 LLGPSSSPFPQHSSKPLSPTQQPPPSSPPFSQLLLLSYQPC----NSLLPWFFPLHHKHN 567
Query: 555 LPNTPPVNVR-PYRYPHSQKEAMATILTDMLQEGIVVP 591
L +P +++ P YP K ++ L LQ + P
Sbjct: 566 L*LSPQHSLQLPLFYPWHHKHSLLYSLQHSLQHPLFSP 453
>TC93746 weakly similar to GP|22830935|dbj|BAC15800. hypothetical
protein~similar to gag-pol polyprotein {Oryza sativa
(japonica cultivar-group)}, partial (4%)
Length = 1019
Score = 33.5 bits (75), Expect = 0.59
Identities = 18/47 (38%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Frame = +2
Query: 1103 LLQPLPTPAQIWEDISMDFITGLPPSHGHTVAWVIV-DRLSKYAHFV 1148
LL+P+P P WED+++DF GL + + ++V D+ S+ AHF+
Sbjct: 464 LLRPVPKPP--WEDVTIDFSLGLL*TQQLKDSKMVVGDKFSRMAHFI 598
>TC88376 similar to GP|8778315|gb|AAF79324.1| F14J16.29 {Arabidopsis
thaliana}, partial (25%)
Length = 706
Score = 32.3 bits (72), Expect = 1.3
Identities = 22/70 (31%), Positives = 30/70 (42%)
Frame = +1
Query: 493 TITFLPSPPSTPSPQFLTLENLSTPPPDFDPALWELLQSYAPVFSTPHGLPPTRFHDHHI 552
T+ LPS P+ P PQ + PP P++ +L P T LP F
Sbjct: 235 TLPPLPSMPTLPQPQG------NVPPLPTIPSMPKLTMPPLPSIPTNPTLPSLNFPPFPS 396
Query: 553 NLLPNTPPVN 562
N LPN P ++
Sbjct: 397 NSLPNIPSIS 426
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.322 0.136 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,261,478
Number of Sequences: 36976
Number of extensions: 810912
Number of successful extensions: 5196
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 4838
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5158
length of query: 1387
length of database: 9,014,727
effective HSP length: 108
effective length of query: 1279
effective length of database: 5,021,319
effective search space: 6422267001
effective search space used: 6422267001
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0007.12


