

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
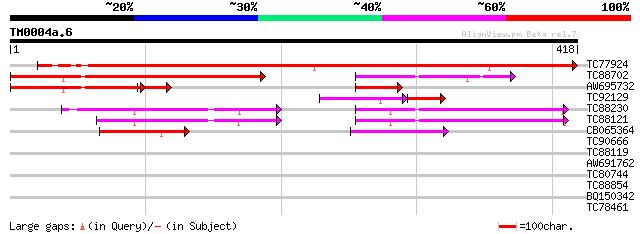
Query= TM0004a.6
(418 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC77924 similar to PIR|H84807|H84807 probable phospholipid cytid... 517 e-147
TC88702 similar to GP|13129435|gb|AAK13093.1 Putative phospholip... 342 1e-94
AW695732 similar to GP|13129435|gb| Putative phospholipid cytidy... 175 5e-47
TC92129 homologue to PIR|H84807|H84807 probable phospholipid cyt... 110 2e-29
TC88230 similar to PIR|T07981|T07981 probable choline-phosphate ... 109 2e-24
TC88121 homologue to PIR|T06558|T06558 choline-phosphate cytidyl... 105 3e-23
CB065364 homologue to PIR|A83022|A83 LPS biosynthesis protein Rf... 60 1e-09
TC90666 weakly similar to GP|21594566|gb|AAM66022.1 unknown {Ara... 30 1.3
TC88119 similar to GP|10178284|emb|CAC08342. rev interacting pro... 29 2.9
AW691762 weakly similar to GP|10177086|db calcium sensor homolog... 29 2.9
TC80744 similar to GP|4587575|gb|AAD25806.1| Belongs to PF|01121... 28 6.6
TC88854 similar to GP|7767667|gb|AAF69164.1| F27F5.26 {Arabidops... 28 6.6
BQ150342 28 8.6
TC78461 weakly similar to GP|22136978|gb|AAM91718.1 unknown prot... 28 8.6
>TC77924 similar to PIR|H84807|H84807 probable phospholipid
cytidylyltransferase [imported] - Arabidopsis thaliana,
partial (86%)
Length = 1691
Score = 517 bits (1332), Expect = e-147
Identities = 255/412 (61%), Positives = 324/412 (77%), Gaps = 14/412 (3%)
Frame = +2
Query: 21 GGLMVTAALLGLSTSYFGGIGHLPLPFPWSNSGIFHKKKSGKRRVRVYMDGCFDLMHYGH 80
G ++V ++LG ++ G L W +G ++ K+ +RVYMDGCFD+MHYGH
Sbjct: 131 GVIIVGVSVLG----FYSSSGPFGL---WGKNG----RRRNKKPIRVYMDGCFDMMHYGH 277
Query: 81 ANALRQAKALGDELVVGLVNDEEILATKGPPVLTMAERLALVSGLKWVDEVITDAPYAIT 140
NALRQA+ALGD+L+VG+V+D+EI+A KGPPV + ERL +V+ +KWVDEVI +APYAIT
Sbjct: 278 CNALRQARALGDQLIVGVVSDDEIIANKGPPVTPLHERLIMVNAVKWVDEVIPEAPYAIT 457
Query: 141 EQFLHRLFHEYKIDYVIHGDDPCLLPDGTDAYAAAKKAGRYKQIKRTEGVSSTDIVGRIM 200
E+F+ +LF EY IDY+IHGDDPC+LPDGTDAYA AKKAGRYKQIKRTEGVSSTDIVGR++
Sbjct: 458 EEFMKKLFDEYNIDYIIHGDDPCVLPDGTDAYAHAKKAGRYKQIKRTEGVSSTDIVGRML 637
Query: 201 SSLKDQKTCEDQNGTDVKRQEES------------MSKDSHISQFLPTSRRIVQFSNGKG 248
++++ + N + ++RQ + ++ + +S FLPTSRRIVQFSNG+
Sbjct: 638 LCVRERNIADTHNHSSLQRQFSNGRGQQKFEDGGVVASGTRVSHFLPTSRRIVQFSNGRS 817
Query: 249 PGPNARIVYIDGAFDLFHAGHVEILKRARELGDFLLVGIHSDETVSEHRGNHYPIMHLHE 308
PGP+ARIVYIDGAFDLFHAGHVEIL+ AR+ GDFLLVGIH+D+TVS RG H PIM LHE
Sbjct: 818 PGPDARIVYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLHRPIMSLHE 997
Query: 309 RSLSVLACRHVDEVIIGAPWEITKDMITTFNISLVVHGTVAEKS--LHSERDPYEVPKSM 366
RSLSVLACR+VDEVIIGAPWE++KDMITTFNISLVVHGT AE + + +PY VP S+
Sbjct: 998 RSLSVLACRYVDEVIIGAPWEVSKDMITTFNISLVVHGTTAENTDFQKEQSNPYAVPISL 1177
Query: 367 GMFRLLESPKDITTTTVAQRILANHEAYEKRNAKKSQSEKRYYEEKNYVSGD 418
G+F++L+SP DITTTT+ +RI++NHEAY+KRN KK +SEK+YYE K +VSGD
Sbjct: 1178GIFQILDSPLDITTTTIIRRIVSNHEAYQKRNEKKGESEKKYYESKTHVSGD 1333
>TC88702 similar to GP|13129435|gb|AAK13093.1 Putative phospholipid
cytidylyltransferase {Oryza sativa}, partial (17%)
Length = 801
Score = 342 bits (878), Expect = 1e-94
Identities = 169/190 (88%), Positives = 175/190 (91%), Gaps = 2/190 (1%)
Frame = +1
Query: 1 MDYESNSWIWEGVYYYPHLFGGLMVTAALLGLSTSYFG--GIGHLPLPFPWSNSGIFHKK 58
MDYESNSWIWEGVYYYPHLFGGLMVTAALLGLSTSYFG G+ LPLP W N G KK
Sbjct: 238 MDYESNSWIWEGVYYYPHLFGGLMVTAALLGLSTSYFGVIGVPSLPLPCSWYNLG--KKK 411
Query: 59 KSGKRRVRVYMDGCFDLMHYGHANALRQAKALGDELVVGLVNDEEILATKGPPVLTMAER 118
K+GKRRVRVYMDGCFDLMHYGHANALRQAKALGDELVVGLV+DEEI+A KGPPVL+M ER
Sbjct: 412 KTGKRRVRVYMDGCFDLMHYGHANALRQAKALGDELVVGLVSDEEIVANKGPPVLSMDER 591
Query: 119 LALVSGLKWVDEVITDAPYAITEQFLHRLFHEYKIDYVIHGDDPCLLPDGTDAYAAAKKA 178
LALVSGLKWVDEVITDAPYAITE FL+RLFHEY IDYVIHGDDPCLLPDGTDAYAAAKKA
Sbjct: 592 LALVSGLKWVDEVITDAPYAITETFLNRLFHEYNIDYVIHGDDPCLLPDGTDAYAAAKKA 771
Query: 179 GRYKQIKRTE 188
GRYKQIKRTE
Sbjct: 772 GRYKQIKRTE 801
Score = 77.0 bits (188), Expect = 1e-14
Identities = 45/122 (36%), Positives = 67/122 (54%), Gaps = 4/122 (3%)
Frame = +1
Query: 256 VYIDGAFDLFHAGHVEILKRARELGDFLLVGIHSDETVSEHRGNHYPIMHLHERSLSVLA 315
VY+DG FDL H GH L++A+ LGD L+VG+ SDE + ++G P++ + ER V
Sbjct: 436 VYMDGCFDLMHYGHANALRQAKALGDELVVGLVSDEEIVANKGP--PVLSMDERLALVSG 609
Query: 316 CRHVDEVIIGAPWEITKDMIT----TFNISLVVHGTVAEKSLHSERDPYEVPKSMGMFRL 371
+ VDEVI AP+ IT+ + +NI V+HG L D Y K G ++
Sbjct: 610 LKWVDEVITDAPYAITETFLNRLFHEYNIDYVIHGD-DPCLLPDGTDAYAAAKKAGRYKQ 786
Query: 372 LE 373
++
Sbjct: 787 IK 792
>AW695732 similar to GP|13129435|gb| Putative phospholipid
cytidylyltransferase {Oryza sativa}, partial (7%)
Length = 497
Score = 175 bits (443), Expect(2) = 5e-47
Identities = 86/102 (84%), Positives = 90/102 (87%), Gaps = 2/102 (1%)
Frame = +3
Query: 1 MDYESNSWIWEGVYYYPHLFGGLMVTAALLGLSTSYFG--GIGHLPLPFPWSNSGIFHKK 58
MDYESNSWIWEGVYYYPHLFGGLMVTAALLGLSTSYFG G+ LPLP W N G KK
Sbjct: 138 MDYESNSWIWEGVYYYPHLFGGLMVTAALLGLSTSYFGVIGVPSLPLPCSWYNLG--KKK 311
Query: 59 KSGKRRVRVYMDGCFDLMHYGHANALRQAKALGDELVVGLVN 100
K+GKRRV VYMDGCFDLMHYGHANAL+QAKALGDELVVGLV+
Sbjct: 312 KTGKRRVHVYMDGCFDLMHYGHANALKQAKALGDELVVGLVS 437
Score = 45.1 bits (105), Expect = 5e-05
Identities = 20/34 (58%), Positives = 25/34 (72%)
Frame = +3
Query: 256 VYIDGAFDLFHAGHVEILKRARELGDFLLVGIHS 289
VY+DG FDL H GH LK+A+ LGD L+VG+ S
Sbjct: 336 VYMDGCFDLMHYGHANALKQAKALGDELVVGLVS 437
Score = 30.8 bits (68), Expect(2) = 5e-47
Identities = 13/25 (52%), Positives = 20/25 (80%)
Frame = +1
Query: 95 VVGLVNDEEILATKGPPVLTMAERL 119
++ L+ DEEI+A KGPPVL+M ++
Sbjct: 421 LLDLLVDEEIVANKGPPVLSMDRKV 495
>TC92129 homologue to PIR|H84807|H84807 probable phospholipid
cytidylyltransferase [imported] - Arabidopsis thaliana,
partial (14%)
Length = 510
Score = 110 bits (274), Expect(2) = 2e-29
Identities = 63/114 (55%), Positives = 65/114 (56%), Gaps = 49/114 (42%)
Frame = +1
Query: 229 HISQFLPTSRRIVQFSNGKGPGPNARIVYIDGAFDLFHAGHVEI---------------- 272
HISQFLPTSRRIVQFSNGKGPGPNARIVYIDGAFDLFHAGHV++
Sbjct: 1 HISQFLPTSRRIVQFSNGKGPGPNARIVYIDGAFDLFHAGHVQV*IGFM*GVVYYPQLNA 180
Query: 273 ---------------------------------LKRARELGDFLLVGIHSDETV 293
LKRARELGDFLLVGIHSDETV
Sbjct: 181 *VHGSP*PNRQEPFDPIILI*LCLIHYFVLYQMLKRARELGDFLLVGIHSDETV 342
Score = 37.0 bits (84), Expect(2) = 2e-29
Identities = 19/29 (65%), Positives = 21/29 (71%), Gaps = 1/29 (3%)
Frame = +2
Query: 294 SEHRGNHYPIMHLHERSL-SVLACRHVDE 321
SE+RGNHYPIMHLHE A R+VDE
Sbjct: 419 SENRGNHYPIMHLHEAEP*RG*ASRYVDE 505
Score = 27.7 bits (60), Expect = 8.6
Identities = 10/14 (71%), Positives = 11/14 (78%)
Frame = +1
Query: 67 VYMDGCFDLMHYGH 80
VY+DG FDL H GH
Sbjct: 82 VYIDGAFDLFHAGH 123
>TC88230 similar to PIR|T07981|T07981 probable choline-phosphate
cytidylyltransferase (EC 2.7.7.15) (clone CCT1) - rape,
partial (59%)
Length = 1364
Score = 109 bits (273), Expect = 2e-24
Identities = 69/166 (41%), Positives = 92/166 (54%), Gaps = 4/166 (2%)
Frame = +3
Query: 39 GIGHLPLPFPWSNSGIFHKKKSGKRRVRVYMDGCFDLMHYGHANALRQAKAL--GDELVV 96
GIG++ S+S I H R VRVY DG +DL H+GHA +L QAK L+V
Sbjct: 57 GIGNMT-----SSSTIPHLPPPEDRPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLV 221
Query: 97 GLVNDEEILATKGPPVLTMAERLALVSGLKWVDEVITDAPYAITEQFLHRLFHEYKIDYV 156
G +D KG V+T ER + KWVDEVI DAP+ I ++FL + KID+V
Sbjct: 222 GCCSDAVTHKYKGKTVMTEDERYESLRHCKWVDEVIPDAPWVINQEFLDK----NKIDFV 389
Query: 157 IHGDDPCLLPDG--TDAYAAAKKAGRYKQIKRTEGVSSTDIVGRIM 200
H P G D Y K GR+K+ +RTEG+S++DI+ RI+
Sbjct: 390 AHDSLPYADTSGAANDVYEFVKAVGRFKETQRTEGISTSDIIMRIV 527
Score = 85.9 bits (211), Expect = 3e-17
Identities = 54/164 (32%), Positives = 86/164 (51%), Gaps = 7/164 (4%)
Frame = +3
Query: 256 VYIDGAFDLFHAGHVEILKRAREL--GDFLLVGIHSDETVSEHRGNHYPIMHLHERSLSV 313
VY DG +DLFH GH L++A++ +LLVG SD +++G +M ER S+
Sbjct: 126 VYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDAVTHKYKGK--TVMTEDERYESL 299
Query: 314 LACRHVDEVIIGAPWEITKDMITTFNISLVVHGTVA-EKSLHSERDPYEVPKSMGMFRLL 372
C+ VDEVI APW I ++ + I V H ++ + + D YE K++G F+
Sbjct: 300 RHCKWVDEVIPDAPWVINQEFLDKNKIDFVAHDSLPYADTSGAANDVYEFVKAVGRFKET 479
Query: 373 ESPKDITTTTVAQRILANHEAYEKRNAKKSQSEK----RYYEEK 412
+ + I+T+ + RI+ ++ Y RN + S K Y +EK
Sbjct: 480 QRTEGISTSDIIMRIVKDYNQYVLRNLDRGYSRKDLGVSYVKEK 611
>TC88121 homologue to PIR|T06558|T06558 choline-phosphate
cytidylyltransferase (EC 2.7.7.15) - garden pea, partial
(97%)
Length = 1063
Score = 105 bits (262), Expect = 3e-23
Identities = 60/140 (42%), Positives = 81/140 (57%), Gaps = 4/140 (2%)
Frame = +3
Query: 65 VRVYMDGCFDLMHYGHANALRQAKAL--GDELVVGLVNDEEILATKGPPVLTMAERLALV 122
VRVY DG +DL H+GHA +L QAK L+VG +DE KG V+ ER +
Sbjct: 141 VRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMNEQERYESL 320
Query: 123 SGLKWVDEVITDAPYAITEQFLHRLFHEYKIDYVIHGDDPCLLPD--GTDAYAAAKKAGR 180
KWVDEVI D P+ I ++F+ + +KIDYV H P G D Y K G+
Sbjct: 321 RHCKWVDEVIPDVPWVINQEFIDK----HKIDYVAHDALPYADTSGAGNDVYEFVKAIGK 488
Query: 181 YKQIKRTEGVSSTDIVGRIM 200
+K+ KRTEG+S++DI+ RI+
Sbjct: 489 FKETKRTEGISTSDIIMRII 548
Score = 89.0 bits (219), Expect = 3e-18
Identities = 55/164 (33%), Positives = 86/164 (51%), Gaps = 7/164 (4%)
Frame = +3
Query: 256 VYIDGAFDLFHAGHVEILKRAREL--GDFLLVGIHSDETVSEHRGNHYPIMHLHERSLSV 313
VY DG +DLFH GH L++A++ +LLVG SDE +++G +M+ ER S+
Sbjct: 147 VYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGK--TVMNEQERYESL 320
Query: 314 LACRHVDEVIIGAPWEITKDMITTFNISLVVHGTVA-EKSLHSERDPYEVPKSMGMFRLL 372
C+ VDEVI PW I ++ I I V H + + + D YE K++G F+
Sbjct: 321 RHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGNDVYEFVKAIGKFKET 500
Query: 373 ESPKDITTTTVAQRILANHEAYEKRNAKKSQSEKR----YYEEK 412
+ + I+T+ + RI+ ++ Y RN + S K Y +EK
Sbjct: 501 KRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEK 632
>CB065364 homologue to PIR|A83022|A83 LPS biosynthesis protein RfaE PA4996
[imported] - Pseudomonas aeruginosa (strain PAO1),
partial (47%)
Length = 682
Score = 60.5 bits (145), Expect = 1e-09
Identities = 29/68 (42%), Positives = 45/68 (65%), Gaps = 2/68 (2%)
Frame = +2
Query: 67 VYMDGCFDLMHYGHANALRQAKALGDELVVGLVNDEEILATKGP--PVLTMAERLALVSG 124
V+ +GCFD++H GH L QA+A GD L+V + +D + KGP P+ ++ R+A+++G
Sbjct: 452 VFTNGCFDILHAGHVTYLEQARAQGDRLIVAVNDDASVSRLKGPGRPINSVDRRMAVLAG 631
Query: 125 LKWVDEVI 132
L VD VI
Sbjct: 632 LGAVDWVI 655
Score = 55.8 bits (133), Expect = 3e-08
Identities = 29/72 (40%), Positives = 42/72 (58%)
Frame = +2
Query: 252 NARIVYIDGAFDLFHAGHVEILKRARELGDFLLVGIHSDETVSEHRGNHYPIMHLHERSL 311
N +IV+ +G FD+ HAGHV L++AR GD L+V ++ D +VS +G PI + R
Sbjct: 440 NEKIVFTNGCFDILHAGHVTYLEQARAQGDRLIVAVNDDASVSRLKGPGRPINSVDRRMA 619
Query: 312 SVLACRHVDEVI 323
+ VD VI
Sbjct: 620 VLAGLGAVDWVI 655
>TC90666 weakly similar to GP|21594566|gb|AAM66022.1 unknown {Arabidopsis
thaliana}, partial (58%)
Length = 664
Score = 30.4 bits (67), Expect = 1.3
Identities = 17/68 (25%), Positives = 36/68 (52%), Gaps = 1/68 (1%)
Frame = +1
Query: 159 GDDPCLLPDGTDAYAAAKKAGRYKQ-IKRTEGVSSTDIVGRIMSSLKDQKTCEDQNGTDV 217
G +P L+ + Y AKK +YK+ +K+ S + R + +L + + +D+NG
Sbjct: 25 GYNPSLIKKQREIYNNAKKVKKYKKMLKQQNHQSDPSVSQRHIENLNETEEDKDKNGRRR 204
Query: 218 KRQEESMS 225
++++ + S
Sbjct: 205 RKKDSAFS 228
>TC88119 similar to GP|10178284|emb|CAC08342. rev interacting protein
mis3-like {Arabidopsis thaliana}, partial (79%)
Length = 1369
Score = 29.3 bits (64), Expect = 2.9
Identities = 14/48 (29%), Positives = 24/48 (49%), Gaps = 3/48 (6%)
Frame = +1
Query: 328 WEITKDMITTFNISL---VVHGTVAEKSLHSERDPYEVPKSMGMFRLL 372
W + K + F IS +V G++ + +DPY + K+ + RLL
Sbjct: 349 WPLVKSSLKEFGISAELNLVEGSMTVSTTRKTKDPYIIVKARDLIRLL 492
>AW691762 weakly similar to GP|10177086|db calcium sensor homolog
{Arabidopsis thaliana}, partial (63%)
Length = 653
Score = 29.3 bits (64), Expect = 2.9
Identities = 15/34 (44%), Positives = 20/34 (58%), Gaps = 3/34 (8%)
Frame = +3
Query: 252 NARIVYIDGAFDLF---HAGHVEILKRARELGDF 282
N R +++D FDLF H GH+E + R LG F
Sbjct: 363 NKRNLFVDRMFDLFDVNHNGHIEFGEFIRSLGIF 464
>TC80744 similar to GP|4587575|gb|AAD25806.1| Belongs to PF|01121
Uncharacterized protein family UPF0038 containing
ATP/GTP binding domain. ESTs, partial (77%)
Length = 1203
Score = 28.1 bits (61), Expect = 6.6
Identities = 19/58 (32%), Positives = 23/58 (38%), Gaps = 1/58 (1%)
Frame = +3
Query: 305 HLHERSLSVLACRHVDEVIIGAPWEITKDMITTFNISLVVHGTVAEKSL-HSERDPYE 361
HL+ L L GAPW + T +L VHG+V S H DP E
Sbjct: 519 HLYRSELDALKNPEEAHARRGAPWTFNPTRLLTCLKNLRVHGSVYAPSFDHGVGDPVE 692
>TC88854 similar to GP|7767667|gb|AAF69164.1| F27F5.26 {Arabidopsis
thaliana}, partial (22%)
Length = 1043
Score = 28.1 bits (61), Expect = 6.6
Identities = 11/15 (73%), Positives = 12/15 (79%)
Frame = -1
Query: 154 DYVIHGDDPCLLPDG 168
D IHGDDP L+PDG
Sbjct: 224 DLSIHGDDPGLVPDG 180
>BQ150342
Length = 603
Score = 27.7 bits (60), Expect = 8.6
Identities = 13/48 (27%), Positives = 24/48 (49%)
Frame = -1
Query: 269 HVEILKRARELGDFLLVGIHSDETVSEHRGNHYPIMHLHERSLSVLAC 316
++E+ +R + LL+G+HS ++ N + + LSVL C
Sbjct: 534 YIELAD*SRSSAEMLLLGLHSQRQLTSVHHNRLD*LRTYIELLSVLVC 391
>TC78461 weakly similar to GP|22136978|gb|AAM91718.1 unknown protein
{Arabidopsis thaliana}, partial (65%)
Length = 1464
Score = 27.7 bits (60), Expect = 8.6
Identities = 16/53 (30%), Positives = 24/53 (45%), Gaps = 10/53 (18%)
Frame = +1
Query: 4 ESNSWIWE----------GVYYYPHLFGGLMVTAALLGLSTSYFGGIGHLPLP 46
E NSWIW V+Y + G+ + L+G ++ G I + PLP
Sbjct: 553 EINSWIWGKDKKIDEKGFNVHYVRPINRGIEAPSVLVGTFLAHVGEITNTPLP 711
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.319 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,545,790
Number of Sequences: 36976
Number of extensions: 174661
Number of successful extensions: 841
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 804
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 825
length of query: 418
length of database: 9,014,727
effective HSP length: 99
effective length of query: 319
effective length of database: 5,354,103
effective search space: 1707958857
effective search space used: 1707958857
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0004a.6


