

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0004a.4
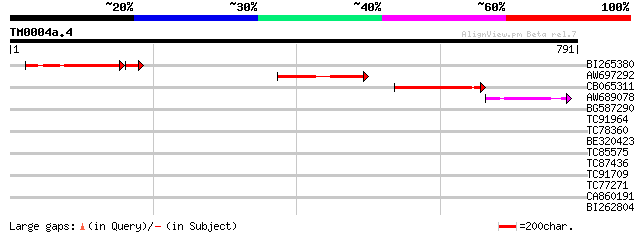
(791 letters)
Database: MTGI
36,976 sequences; 27,044,181 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI265380 weakly similar to GP|17932898|em isoamylase {Triticum a... 162 8e-48
AW697292 homologue to GP|22136708|gb putative isoamylase {Arabid... 150 2e-36
CB065311 similar to PIR|C83375|C83 probable glycosyl hydrolase P... 141 1e-33
AW689078 similar to GP|22136708|gb putative isoamylase {Arabidop... 49 6e-06
BG587290 33 0.42
TC91964 similar to GP|23496165|gb|AAN35825.1 hypothetical protei... 32 0.95
TC78360 30 2.8
BE320423 30 4.7
TC85575 similar to PIR|D86410|D86410 protein F3M18.16 [imported]... 29 6.1
TC87436 similar to PIR|T51575|T51575 2-hydroxyphytanoyl-CoA lyas... 29 8.0
TC91709 homologue to GP|21213868|emb|CAD12767. LHY protein {Phas... 29 8.0
TC77271 similar to PIR|T07929|T07929 probable long-chain-fatty-a... 29 8.0
CA860191 homologue to GP|19697342|gb hypothetical protein {Dicty... 29 8.0
BI262804 weakly similar to GP|11072027|gb F12A21.14 {Arabidopsis... 29 8.0
>BI265380 weakly similar to GP|17932898|em isoamylase {Triticum aestivum},
partial (13%)
Length = 604
Score = 162 bits (409), Expect(2) = 8e-48
Identities = 83/140 (59%), Positives = 106/140 (75%), Gaps = 1/140 (0%)
Frame = +3
Query: 22 HSDSQCRVSLSKRVSEKHKSICSTTKILATGNGSGFETETTLV-VDKPQLGGRFQVSRGY 80
H+ + +S + R +K K KI A GN G +TET ++ ++K QL Q+S+G+
Sbjct: 138 HNRTSIIISPNSRPKKKKK------KIFAIGNRVGVDTETAVIEIEKRQL----QLSKGH 287
Query: 81 PAPFGATVRDGGVNFAIYSLNAVSATLCLFTLSDFQDNQVTEYITLDPLMNKTGSVWHVF 140
P+P+GAT ++ GVNFAI SLN++SATLC FTLSDF++N+VTEYITLDPL+NKTG VWHVF
Sbjct: 288 PSPYGATPQEDGVNFAINSLNSLSATLCFFTLSDFKNNKVTEYITLDPLVNKTGCVWHVF 467
Query: 141 LKGDFGDMLYGYKFDGKFSP 160
LKGDF DMLY YKFDGKFSP
Sbjct: 468 LKGDFKDMLYAYKFDGKFSP 527
Score = 47.8 bits (112), Expect(2) = 8e-48
Identities = 21/25 (84%), Positives = 23/25 (92%)
Frame = +2
Query: 162 EGHYYDSSLILLDPYAKAVISRGEF 186
+GHYYDSS IL+DPYAKAVISR EF
Sbjct: 530 QGHYYDSSRILIDPYAKAVISRXEF 604
>AW697292 homologue to GP|22136708|gb putative isoamylase {Arabidopsis
thaliana}, partial (13%)
Length = 343
Score = 150 bits (379), Expect = 2e-36
Identities = 70/127 (55%), Positives = 85/127 (66%)
Frame = +2
Query: 374 KGEFYNYSGCGNTLNCNHPVVRQFIVDCLRYWVTEMHVDGFRFDLASIMTRSSSLWNGVN 433
KG+ N+SGCGNTLNCNHPVV I+D LR+WVTE HVDGFRFDLASI+ R +
Sbjct: 5 KGQLLNFSGCGNTLNCNHPVVMDLILDSLRHWVTEYHVDGFRFDLASILCRGTD------ 166
Query: 434 VFGTSIEGDLLATGTPLVSPPLIDLISNDPILHGVKLIAEAWDAGGLYQVGTFPHWGIWS 493
G+PL +PPLI I+ D +L K+IAE WD GGLY VG+FP+W W+
Sbjct: 167 -------------GSPLNAPPLIRAIAKDAVLSRCKIIAEPWDCGGLYLVGSFPNWDRWA 307
Query: 494 EWNGKYR 500
EWNGKY+
Sbjct: 308 EWNGKYQ 328
>CB065311 similar to PIR|C83375|C83 probable glycosyl hydrolase PA2160
[imported] - Pseudomonas aeruginosa (strain PAO1),
partial (26%)
Length = 567
Score = 141 bits (355), Expect = 1e-33
Identities = 62/128 (48%), Positives = 85/128 (65%)
Frame = -2
Query: 537 NSINFVCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFASSSVKKLRK 596
+S+NF+ HDGFTL DLV+YN+KHN N E+N DG ++N SWNCG EG + LR
Sbjct: 566 SSVNFITAHDGFTLRDLVSYNHKHNEDNDENNQDGTDNNLSWNCGAEGPTDDPDINALRM 387
Query: 597 RQMRNFFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYLNYFQWDIKEESSSDFFRF 656
RQMRNFF +L+ +QG PMI GDE+ T+ GNNN YC D+ + + W++ +E +D F
Sbjct: 386 RQMRNFFATLLFAQGTPMIVAGDEFSRTQHGNNNAYCQDSEIGWINWEL-DEDGADLLAF 210
Query: 657 CCLMTKFR 664
+T+ R
Sbjct: 209 VKRLTRLR 186
>AW689078 similar to GP|22136708|gb putative isoamylase {Arabidopsis
thaliana}, partial (13%)
Length = 511
Score = 49.3 bits (116), Expect = 6e-06
Identities = 35/122 (28%), Positives = 57/122 (46%), Gaps = 2/122 (1%)
Frame = +3
Query: 664 RYECESLGLDDFPTSDRLQWHGHFPGKPDWSE-TSRFVAFTMVDSVKREIYIAFNTSHLP 722
R+ ++L ++F + + + WH + +W S+F+AFT+ D +IY+AFN
Sbjct: 42 RHAHKTLSHENFLSENEITWH-----EDNWDNYESKFLAFTLHDKSGGDIYLAFNAHDYF 206
Query: 723 VTITLPERPGYR-WEPLVDTGKSAPYDFLTPDLPGRDIAIQQYAQFLDANLYPMLSYSSI 781
+ LP P R W +VDT +P D + +PG Y + YSSI
Sbjct: 207 LKALLPTPPTKRKWFRVVDTNLQSPDDIVLDGVPG------------IGETYSIAPYSSI 350
Query: 782 IL 783
+L
Sbjct: 351 LL 356
>BG587290
Length = 667
Score = 33.1 bits (74), Expect = 0.42
Identities = 20/60 (33%), Positives = 33/60 (54%), Gaps = 1/60 (1%)
Frame = +3
Query: 495 WNGKYRDTVRQFVKGTDGFAGAFAECLCGSPNVYQGGGRKPWNS-INFVCTHDGFTLADL 553
W+ +YRD++ F+ G F GA+ + + + Q GGR PW++ I + H + L DL
Sbjct: 378 WSQQYRDSIVNFIFGLR-FKGAYFPWVLLAFDSLQMGGRIPWDTLIGIITGHIFYFLYDL 554
>TC91964 similar to GP|23496165|gb|AAN35825.1 hypothetical protein
{Plasmodium falciparum 3D7}, partial (1%)
Length = 827
Score = 32.0 bits (71), Expect = 0.95
Identities = 19/49 (38%), Positives = 25/49 (50%)
Frame = +1
Query: 540 NFVCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCGQEGEFAS 588
N + T D + D NN +N N + NND +N NN+ N G E AS
Sbjct: 370 NVIGTVDIVSTTDFSENNNDNN-NNNDKNNDNDNENNNDNGGGGNESAS 513
>TC78360
Length = 805
Score = 30.4 bits (67), Expect = 2.8
Identities = 16/59 (27%), Positives = 29/59 (49%)
Frame = +1
Query: 523 GSPNVYQGGGRKPWNSINFVCTHDGFTLADLVTYNNKHNLPNGEDNNDGENHNNSWNCG 581
GSP++ QG + W HD +T D+ + + + +G ++ G N+NN + G
Sbjct: 142 GSPHLIQGQDQHVWEE---KVKHDPYTSHDMGVASRETKVKSGNGDHWGSNYNNGVSNG 309
>BE320423
Length = 210
Score = 29.6 bits (65), Expect = 4.7
Identities = 23/72 (31%), Positives = 27/72 (36%), Gaps = 11/72 (15%)
Frame = +2
Query: 513 FAGAFAECLCGSPNVYQGGGRKPWNSINFVCTHDGFTLADLVTYNNKHN----------- 561
F G G PN YQ G RKP N THDG+ ++N +
Sbjct: 2 FGGDHRATSVGDPNPYQEG-RKPIGGGNLRSTHDGY-------HDNGYGGDKEGRKPVGI 157
Query: 562 LPNGEDNNDGEN 573
P G N DG N
Sbjct: 158 SPTGNSNYDGPN 193
>TC85575 similar to PIR|D86410|D86410 protein F3M18.16 [imported] -
Arabidopsis thaliana, partial (15%)
Length = 1225
Score = 29.3 bits (64), Expect = 6.1
Identities = 23/121 (19%), Positives = 43/121 (35%), Gaps = 5/121 (4%)
Frame = +3
Query: 524 SPNVYQGGGRKPWNSINFVCTHDGFTLADLVT--YNNKHNLPNGEDNNDGENHNNSWNCG 581
+P Y +++ + + D F+ YN+ N N ++N +NN+ +
Sbjct: 450 TPTSYSSSSNNNYDNNKYSSSSDDFSNKKFPEEGYNSNENQNNNNNDNKYFYNNNNKDSS 629
Query: 582 QEGEFASSSVKKLRKRQMRN---FFLSLMVSQGVPMIYMGDEYGHTKGGNNNTYCHDNYL 638
+F + R N F+ + + + Y + NNN Y NY
Sbjct: 630 SNTKFTEEGYNSMENRNNNNNKYFYNNNNKDSSSNTKFPEEGYNSMQNQNNNNYEKYNYN 809
Query: 639 N 639
N
Sbjct: 810 N 812
>TC87436 similar to PIR|T51575|T51575 2-hydroxyphytanoyl-CoA lyase-like
protein - Arabidopsis thaliana, partial (75%)
Length = 1592
Score = 28.9 bits (63), Expect = 8.0
Identities = 15/33 (45%), Positives = 19/33 (57%)
Frame = +1
Query: 680 RLQWHGHFPGKPDWSETSRFVAFTMVDSVKREI 712
RL W HF +P WS+ +FV +VD K EI
Sbjct: 1015 RLNWLLHFGEEPKWSKDVKFV---LVDVSKEEI 1104
>TC91709 homologue to GP|21213868|emb|CAD12767. LHY protein {Phaseolus
vulgaris}, partial (8%)
Length = 649
Score = 28.9 bits (63), Expect = 8.0
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = -2
Query: 564 NGEDNNDGENHNNSWNCGQEGEFASSSVKKL 594
N +NN+ N+N WNC SSS KK+
Sbjct: 276 NNNNNNNNNNNNKDWNC-------SSSSKKI 205
>TC77271 similar to PIR|T07929|T07929 probable long-chain-fatty-acid--CoA
ligase (EC 6.2.1.3) isoform 2 - rape, partial (94%)
Length = 2486
Score = 28.9 bits (63), Expect = 8.0
Identities = 15/59 (25%), Positives = 29/59 (48%)
Frame = -3
Query: 291 RVNFWGYSTVNYFSPMIRYSSAGIQNCGRDGINEVKFLIKEAHKRGIEVIMDVVFNHTV 349
R+ GY+ +F PM + AG+ N ++G+ V ++R + + + VFN +
Sbjct: 1917 RIEVHGYTI--FFCPMFKCWFAGVNNSNKEGLKTVSVNPYRFNRRNLAINILQVFNGNI 1747
>CA860191 homologue to GP|19697342|gb hypothetical protein {Dictyostelium
discoideum}, partial (1%)
Length = 504
Score = 28.9 bits (63), Expect = 8.0
Identities = 10/23 (43%), Positives = 16/23 (69%)
Frame = +2
Query: 557 NNKHNLPNGEDNNDGENHNNSWN 579
NN +N N +NN+ +NH+N +N
Sbjct: 311 NNNNNNSNNNNNNNEDNHHNKFN 379
>BI262804 weakly similar to GP|11072027|gb F12A21.14 {Arabidopsis thaliana},
partial (7%)
Length = 653
Score = 28.9 bits (63), Expect = 8.0
Identities = 16/44 (36%), Positives = 19/44 (42%)
Frame = -3
Query: 114 DFQDNQVTEYITLDPLMNKTGSVWHVFLKGDFGDMLYGYKFDGK 157
+ D YITL L T + KG FG + YG DGK
Sbjct: 270 NLMDEGTAYYITLSDLKVATNNFSKKIGKGSFGSVYYGKMKDGK 139
Database: MTGI
Posted date: Oct 22, 2004 3:39 PM
Number of letters in database: 27,044,181
Number of sequences in database: 36,976
Lambda K H
0.320 0.139 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,275,785
Number of Sequences: 36976
Number of extensions: 454888
Number of successful extensions: 2190
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 2117
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2172
length of query: 791
length of database: 9,014,727
effective HSP length: 104
effective length of query: 687
effective length of database: 5,169,223
effective search space: 3551256201
effective search space used: 3551256201
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0004a.4


