

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0588b.3
(432 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
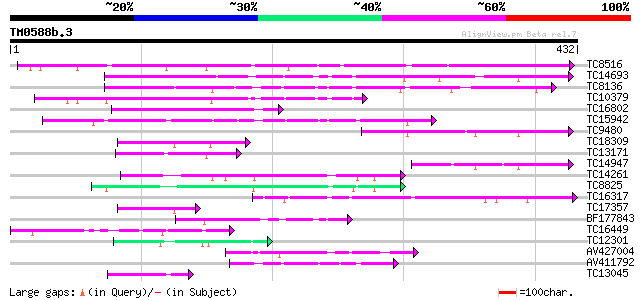
Score E
Sequences producing significant alignments: (bits) Value
TC8516 similar to UP|AAP72988 (AAP72988) CDR1, partial (27%) 244 2e-65
TC14693 similar to GB|BAB01116.1|11994113|AB026658 CND41, chloro... 205 1e-53
TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabid... 129 9e-31
TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast ... 125 1e-29
TC16802 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Ar... 115 1e-26
TC15942 similar to UP|Q9FL43 (Q9FL43) Nucleoid DNA-binding-like ... 101 3e-22
TC9480 weakly similar to UP|Q9AWT3 (Q9AWT3) Nucleoid DNA-binding... 75 3e-14
TC18309 similar to UP|Q9FG60 (Q9FG60) Aspartyl protease-like, pa... 75 3e-14
TC13171 70 5e-13
TC14947 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Ar... 65 2e-11
TC14261 weakly similar to UP|Q05929 (Q05929) EDGP precursor (Fra... 64 4e-11
TC8825 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific ... 62 2e-10
TC16317 similar to UP|Q8W4C5 (Q8W4C5) Nucellin-like protein, par... 60 5e-10
TC17357 similar to UP|Q9FFC3 (Q9FFC3) Protease-like protein, par... 59 2e-09
BF177843 54 4e-08
TC16449 similar to UP|Q940R4 (Q940R4) AT4g16560/dl4305c, partial... 50 9e-07
TC12301 weakly similar to UP|CAR5_RHINI (P43232) Rhizopuspepsin ... 50 9e-07
AV427004 48 3e-06
AV411792 46 1e-05
TC13045 weakly similar to UP|Q9FL43 (Q9FL43) Nucleoid DNA-bindin... 45 3e-05
>TC8516 similar to UP|AAP72988 (AAP72988) CDR1, partial (27%)
Length = 1603
Score = 244 bits (624), Expect = 2e-65
Identities = 154/460 (33%), Positives = 237/460 (51%), Gaps = 36/460 (7%)
Frame = +1
Query: 7 FFHLCFLV-----VLYLLSCQ-------TPIEAQDAGFSVQLTRQNSPHSPFYKPD---- 50
FFHL FL +L SC T + F++ L +SP SPFY
Sbjct: 40 FFHLHFLKHACFSILLAASCSLLATLPFTEPSKTPSSFTIDLIHHDSPLSPFYNSSMTRS 219
Query: 51 ------------NLHRHKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLGSPPVDI 98
++ L H + + + P + NNG+YLM++ +G+P V+
Sbjct: 220 QLIRNAAMRSISRANQLSLSLSHSLNQLKESSPEPI---IIPNNGNYLMRIYIGTPSVER 390
Query: 99 YGLVDTGSDLVWAQCSPCHG--CYKQKSPMFEPLSSKTFNPIPCDSEQCGSL--FSHSCS 154
+ DTGSDL W QCSPC C+ Q +P+++PL+S TF +PCDS+ C L + CS
Sbjct: 391 LAIADTGSDLTWVQCSPCDNTKCFAQNTPLYDPLNSSTFTLLPCDSQPCTQLPYSQYVCS 570
Query: 155 PQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNEN--D 212
C Y+Y+Y D+S + G L+ ++I F + + +FGCG N +++
Sbjct: 571 DYGDCIYAYTYGDNSYSYGGLSSDSIRFDA--TAATPTIPKFVFGCGFQNKFTADKSGKT 744
Query: 213 MGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTP 272
G++GLG GPLSLVSQ+G G +FS CL+PF ++S + + FG+A+ V G GVV+TP
Sbjct: 745 TGIVGLGAGPLSLVSQLGDEIG-HKFSYCLLPFSSNSNSK--LKFGEAAIVQGNGVVSTP 915
Query: 273 LVSEEGQTPYLVTLEGISVGDTFVSFNSSEKLSKGNMMIDSGTPATYLPQEFYDRLVEEL 332
L+ + Y + LEGI+VG V ++ GN++IDSG+ TYL + FY+ V +
Sbjct: 916 LIIKPDLPFYYLNLEGITVGAKTVKTGQTD----GNIIIDSGSTLTYLEESFYNEFVSLV 1083
Query: 333 KVQSSLLPVDNDPDLGTQLCYRSETNLE-GPILTAHFEGADVQLMPIQTFIPPKDGVFCF 391
K ++ + D C+ + + P + HF G DV L P+ T + +D + C
Sbjct: 1084K-ETVAVEEDQYIPYPFDFCFTYKEGMSTPPDVVFHFTGGDVVLKPMNTLVLIEDNLICS 1260
Query: 392 AMAGT-ADGDYIFGNFAQSNILIGFDLDRKTISFKPTDCT 430
+ + DG IFGN Q + +G+D+ +SF PTDC+
Sbjct: 1261TVVPSHFDGIAIFGNLGQIDFHVGYDIQGGKVSFAPTDCS 1380
>TC14693 similar to GB|BAB01116.1|11994113|AB026658 CND41, chloroplast
nucleoid DNA binding protein-like {Arabidopsis
thaliana;} , partial (58%)
Length = 2067
Score = 205 bits (521), Expect = 1e-53
Identities = 129/370 (34%), Positives = 179/370 (47%), Gaps = 13/370 (3%)
Frame = +1
Query: 73 PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSS 132
P S+ +G+Y ++ +G P Y ++DTGSD+ W QC PC CY+Q P+F+P +S
Sbjct: 475 PVSSGTGQGSGEYFSRVGVGQPAKPFYMVLDTGSDVNWLQCKPCADCYQQSDPIFDPTTS 654
Query: 133 KTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELV 192
T+ P+ CD++QC +L +C K C Y SY D S T G ET++F G
Sbjct: 655 STYGPLTCDAQQCQALEVSACRNGK-CLYQVSYGDGSFTVGEFVTETMSF-----GGSGS 816
Query: 193 VGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTS 252
V + GCGH N G F G++GLGGGPLSL SQ+ A FS CLV DS S
Sbjct: 817 VNRVAIGCGHDNEGLF-VGAAGLLGLGGGPLSLTSQIKA----SSFSYCLV--DRDSGKS 975
Query: 253 GTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFN----SSEKLSKGN 308
T+ F A G+ V L +++ T Y V L G+SVG V ++ G
Sbjct: 976 STLEFNSAR--PGDSVTAPLLKNQKIGTFYYVELTGVSVGGQMVGIPPETFEMDQTGAGG 1149
Query: 309 MMIDSGTPATYLPQEFYD-------RLVEELKVQSSLLPVDNDPDLGTQLCYRSETNLEG 361
+++DSGT T L + Y+ RL + L+ + D DL T R
Sbjct: 1150IIVDSGTAITRLRTQAYNSVRDAFRRLTQNLRSAEGVALFDTCYDLSTLQSVRV------ 1311
Query: 362 PILTAHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLDR 419
P ++ HF G +P + ++ P D G FCFA A T I GN Q + FDL +
Sbjct: 1312PTVSLHFSGGKTWSLPAKNYLIPVDSAGTFCFAFAPTTSSLSIIGNVQQQGTRVSFDLAK 1491
Query: 420 KTISFKPTDC 429
+ F P C
Sbjct: 1492NEVGFSPNKC 1521
>TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabidopsis
thaliana;}, partial (70%)
Length = 1659
Score = 129 bits (324), Expect = 9e-31
Identities = 109/364 (29%), Positives = 174/364 (46%), Gaps = 20/364 (5%)
Frame = +1
Query: 73 PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSS 132
P ++ N G+Y++++ +G+P ++ ++DT +D + CS C GC +P F P +S
Sbjct: 322 PIASGQAFNIGNYIVRVKIGTPGQLLFMVLDTSTDEAFVPCSGCTGCSTTTAP-FSPKAS 498
Query: 133 KTFNPIPCDSEQCGSLFSHSC--SPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDE 190
T++P+ C CG + SC + C+++ SYA S+ + L +++++ ++
Sbjct: 499 TTYSPLDCSVPLCGQVRGLSCPATGSATCSFNQSYAGSTFS-ATLVQDSLSLATD----- 660
Query: 191 LVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSR 250
V + FGC ++ SGA G++GLG GPLSL+SQ G Y S FS CL P
Sbjct: 661 -AVPNYSFGCINAISGA-TVPAQGLLGLGRGPLSLLSQTGTNY-SGVFSYCL-PSFKSYY 828
Query: 251 TSGTISFGDASDVSGEGVVTTPLV-SEEGQTPYLVTLEGISVGDTFV-------SFNSSE 302
SG++ G + + TTPL+ + + Y V L GISVG V +FN S
Sbjct: 829 FSGSLKLGPVG--QPKSIRTTPLLRNPHRPSLYYVNLTGISVGRVLVPVPAESLAFNPS- 999
Query: 303 KLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQ-----SSLLPVDNDPDLGTQLCYRSET 357
+ +IDSGT T + Y + EE + Q SSL D C+
Sbjct: 1000--TGAGTVIDSGTVITRFIEPVYAAVREEFRKQVTGPFSSLGAFDT--------CFVKTY 1149
Query: 358 NLEGPILTAHFEGADVQLMPIQTFIPPKDG-VFCFAMAGTADGD----YIFGNFAQSNIL 412
P++T H EG D++L + I G + C AMA + + N+ Q N+
Sbjct: 1150ETLAPVVTLHLEGLDLKLPLENSLIHSSSGSLACLAMAAAPENVNSVLNVIANYQQQNLR 1329
Query: 413 IGFD 416
+ FD
Sbjct: 1330VLFD 1341
>TC10379 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kDa chloroplast nucleoid
DNA binding protein (CND41), partial (43%)
Length = 1002
Score = 125 bits (314), Expect = 1e-29
Identities = 89/279 (31%), Positives = 134/279 (47%), Gaps = 26/279 (9%)
Frame = +3
Query: 20 SCQTPIEAQDAGFSVQLTRQNSP----------HSPFYKPD----NLHRHKLPSFHQVPK 65
SC + E S+++ ++ P HS D N KL + + K
Sbjct: 177 SCSSSSEGPKRKASLEVVHKHGPCSQVKVDVPTHSDILSQDKKRVNYIHSKLSTNYNSAK 356
Query: 66 KAFAPNG-----PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPC-HGC 119
++++ P + +G+Y + + LG+P D+ + DTGSDL W QC PC C
Sbjct: 357 ESYSGQSGGATLPVKSGSLIGSGNYFVVVGLGTPKSDLSLVFDTGSDLTWTQCQPCARSC 536
Query: 120 YKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHS------CSPQKLCAYSYSYADSSVTKG 173
YKQ+ P+F+P S ++ I C S C L S + + K C Y Y DSS + G
Sbjct: 537 YKQQDPIFDPKRSTSYYNITCTSSDCTQLTSATGNDPACATITKACVYGIQYGDSSFSVG 716
Query: 174 VLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALY 233
+RE +T +PT+ VV + +FGCG +N G FN G++GLG P+S V Q Y
Sbjct: 717 FFSRERLTV-TPTD----VVDNFLFGCGQNNQGLFN-GAAGLLGLGRHPISFVQQTSQKY 878
Query: 234 GSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTP 272
++FS CL + + G +SFG A+ V+ V TP
Sbjct: 879 -QKKFSYCL---PSTTSAVGHLSFGSAA-VASRYVKYTP 980
>TC16802 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Arabidopsis
thaliana;}, partial (28%)
Length = 812
Score = 115 bits (288), Expect = 1e-26
Identities = 52/131 (39%), Positives = 75/131 (56%)
Frame = +3
Query: 78 VTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNP 137
+ +G+Y ++ +G+P +Y ++DTGSD+VW QC+PC CY Q P+F+P S+T+
Sbjct: 429 LAQGSGEYFTRIGVGTPARYVYMVLDTGSDVVWLQCAPCRKCYSQADPVFDPSKSRTYAG 608
Query: 138 IPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDII 197
IPC + C L S C+ +K+C Y SY D S T G + ET+TF V +
Sbjct: 609 IPCGAPLCRRLDSAGCNSKKVCQYQVSYGDGSFTFGDFSTETLTFRRSR------VTRVA 770
Query: 198 FGCGHSNSGAF 208
GCGH N G F
Sbjct: 771 LGCGHDNEGLF 803
>TC15942 similar to UP|Q9FL43 (Q9FL43) Nucleoid DNA-binding-like protein,
partial (66%)
Length = 973
Score = 101 bits (251), Expect = 3e-22
Identities = 91/318 (28%), Positives = 139/318 (43%), Gaps = 18/318 (5%)
Frame = +1
Query: 26 EAQDAGFSVQLTRQNSPHSPFYKPDNLHRHKLPSFHQ-------------VPKKAFAPNG 72
++ D G ++Q+ SP SPF P L + Q V ++ P
Sbjct: 73 DSHDHGSTLQVFHVFSPCSPFRPPKPLSWEESVLQMQAKDITRVQFLVSLVAGRSIVPIA 252
Query: 73 PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSS 132
S R + Y+++ +G+PP + +DT +D W CS C GC S +F P S
Sbjct: 253 --SGRQIIQSPTYIVRAKIGNPPQTLLMAMDTSNDAAWVPCSGCDGC---SSALFAPEKS 417
Query: 133 KTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTKGVLARETITFSSPTNGDELV 192
TF + C + +C + + SC C ++++Y SS+ LA++T+T ++
Sbjct: 418 TTFKSVGCAAAECKQVPNPSCG-VSACNFNFTYGSSSIAAN-LAQDTVTLATDP------ 573
Query: 193 VGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQCLVPFHADSRTS 252
V FGC +G+ N G++GLG GPLSL+SQ LY S FS CL P S
Sbjct: 574 VPSYTFGCVAKTTGSSNP-PQGLLGLGRGPLSLLSQTQNLYKS-TFSYCL-PSFKSLNFS 744
Query: 253 GTISFGDASDVSGEGVVTTPLVSEEGQ-TPYLVTLEGISVGDTFVSFNSS----EKLSKG 307
G++ G + + TPL+ + + Y V L I VG V + +
Sbjct: 745 GSLRLGPVAQPL--RIKFTPLLKNPRRPSLYYVNLVAIRVGRKIVDIPPAALAFNPTTGA 918
Query: 308 NMMIDSGTPATYLPQEFY 325
+ DSGT T L Y
Sbjct: 919 GTIFDSGTVYTRLAAPAY 972
>TC9480 weakly similar to UP|Q9AWT3 (Q9AWT3) Nucleoid DNA-binding protein
cnd41-like protein, partial (33%)
Length = 716
Score = 74.7 bits (182), Expect = 3e-14
Identities = 53/170 (31%), Positives = 78/170 (45%), Gaps = 9/170 (5%)
Frame = +1
Query: 269 VTTPLV-SEEGQTPYLVTLEGISVGDTFVSFNSS----EKLSKGNMMIDSGTPATYLPQE 323
VT PL + + T Y + L G+SVG + + + + +G +++DSGT T L E
Sbjct: 10 VTAPLRRNPDLDTFYYLGLTGLSVGGEVLPIPEASFEVDAVGEGGIIVDSGTAVTRLRSE 189
Query: 324 FYDRLVEELKVQSSLLPVDNDPDLGTQLCY--RSETNLEGPILTAHFEGADVQLMPIQTF 381
YD L + K + LP L CY S+T++E P + HF +P + +
Sbjct: 190 VYDELRDAFKRGARGLPAAEGVSL-FDTCYDLSSKTSVEVPTVALHFPDGKELPLPAKNY 366
Query: 382 IPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKTISFKPTDC 429
+ P D G FCFA A T I GN Q +GFD+ + F C
Sbjct: 367 LIPVDSVGTFCFAFAPTTSSLSIIGNVQQQGTRVGFDIANSLVGFSAHSC 516
>TC18309 similar to UP|Q9FG60 (Q9FG60) Aspartyl protease-like, partial (14%)
Length = 624
Score = 74.7 bits (182), Expect = 3e-14
Identities = 38/109 (34%), Positives = 54/109 (48%), Gaps = 8/109 (7%)
Frame = +2
Query: 83 GDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKS-----PMFEPLSSKTFNP 137
G Y K+ LGSP D Y VDTGSD++W C C C ++ +++P SKT
Sbjct: 248 GLYFTKIGLGSPSKDYYVQVDTGSDILWVNCVECTRCPRKSDIGIGLTLYDPKRSKTSEF 427
Query: 138 IPCDSEQCGSLFSH---SCSPQKLCAYSYSYADSSVTKGVLARETITFS 183
+ C+ C S + C + C YS SY D S T G ++ +TF+
Sbjct: 428 VSCEHNFCSSTYEGRILGCKAENPCPYSISYGDGSATTGYYVQDYLTFN 574
>TC13171
Length = 598
Score = 70.5 bits (171), Expect = 5e-13
Identities = 39/101 (38%), Positives = 54/101 (52%), Gaps = 5/101 (4%)
Frame = +3
Query: 81 NNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPC 140
+N + LT+GSPP ++ ++DTGS+L W C S +F P S ++NP PC
Sbjct: 297 HNVSLTVTLTVGSPPQNVTMVLDTGSELSWLHCKKAPN--SNSSSIFNPPLSSSYNPTPC 470
Query: 141 DSEQCGSL-----FSHSCSPQKLCAYSYSYADSSVTKGVLA 176
S C + SC P+KLC + SYADSS +G LA
Sbjct: 471 TSPVCTTRTRDFPLPVSCDPRKLCHATLSYADSSSVEGNLA 593
>TC14947 similar to GB|AAP31963.1|30387595|BT006619 At1g01300 {Arabidopsis
thaliana;}, partial (26%)
Length = 728
Score = 65.5 bits (158), Expect = 2e-11
Identities = 43/128 (33%), Positives = 62/128 (47%), Gaps = 5/128 (3%)
Frame = +2
Query: 307 GNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGT-QLCY--RSETNLEGPI 363
G ++IDSGT T L + Y L + ++ + L + P+ C+ +T ++ P
Sbjct: 8 GGVIIDSGTSVTRLTRPGYIALRDAFRLGA--LHLKRAPEFSLFDTCFDLSGQTEVKVPT 181
Query: 364 LTAHFEGADVQLMPIQTFIPPKD--GVFCFAMAGTADGDYIFGNFAQSNILIGFDLDRKT 421
+ HF GADV L P ++ P D G FCFA AGT G I GN Q + +DL
Sbjct: 182 VVLHFRGADVSL-PATNYLIPVDSSGSFCFAFAGTMSGLSIIGNIQQQGFRVVYDLGSSR 358
Query: 422 ISFKPTDC 429
+ F P C
Sbjct: 359 VGFAPRGC 382
>TC14261 weakly similar to UP|Q05929 (Q05929) EDGP precursor (Fragment),
partial (84%)
Length = 1604
Score = 64.3 bits (155), Expect = 4e-11
Identities = 62/250 (24%), Positives = 105/250 (41%), Gaps = 33/250 (13%)
Frame = +1
Query: 85 YLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQ 144
YL ++ +P V + +VD G + +W C + +S T+ P C S Q
Sbjct: 175 YLTQINQRTPQVPVKLVVDLGGNFLWVDCETNY-------------TSSTYRPARCKSAQ 315
Query: 145 CGSLFSHSC-----SPQKLCAYSY------SYADSSVTKGVLARETITFSSP---TNGDE 190
C S SC SP+ C + + + T G LA++T++ S T G
Sbjct: 316 CSLAGSSSCGDCFSSPKPGCNNNTCGLIPDNTITGTATSGELAQDTLSIQSTNGLTPGRI 495
Query: 191 LVVGDIIFGCGHS-NSGAFNENDMGVIGLGGGPLSLVSQMGALYG-SRRFSQCLVPFHAD 248
V + +F CG + + G+ GLG ++L SQ + + R+F+ CL
Sbjct: 496 ATVSNFLFTCGSTFLLKGLAKGTTGMAGLGRTKIALPSQFSSAFSFHRKFAICL------ 657
Query: 249 SRTSGTISFGDASDV------SGEGVVTTPLVSE-----------EGQTPYLVTLEGISV 291
S ++G + FGD V + +G+ TPL++ E Y + ++ I +
Sbjct: 658 SSSTGVVIFGDGPYVLLPNVDASKGLSFTPLLTNPVSTSSAFSQGEPSAEYFIGVKSIVI 837
Query: 292 GDTFVSFNSS 301
+ V+ NSS
Sbjct: 838 DEKTVTVNSS 867
>TC8825 weakly similar to UP|Q8GT67 (Q8GT67) Xyloglucan-specific fungal
endoglucanase inhibitor protein precursor, partial (49%)
Length = 882
Score = 62.0 bits (149), Expect = 2e-10
Identities = 65/264 (24%), Positives = 107/264 (39%), Gaps = 25/264 (9%)
Frame = +3
Query: 63 VPKKAFAPNG---PFSTRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGC 119
+ + +F P P + VTS+ Y+ ++ +P V + +D G +W C
Sbjct: 105 IAQTSFRPKALVLPITKDVTSSLPQYITQIKQRTPLVPVKLTLDLGGGYLWVNCEN---- 272
Query: 120 YKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCSPQKLCAYSYSYADSSVTK-GVLARE 178
S TF P C S QC CS K+C S S + V+ G + +
Sbjct: 273 --------RQYVSSTFKPARCGSSQCSLFGLTGCSGDKICGRSPSNTVTGVSSYGDIHSD 428
Query: 179 TITFSS---PTNGDELVVGDIIFGCGHS-NSGAFNENDMGVIGLGGGPLSLVSQMGALYG 234
++ +S T + V + +F CG + G+ GLG +SL SQ + +
Sbjct: 429 VVSVNSTDGTTPTKVVSVPNFLFICGSKVVQNGLAKGVTGMAGLGRTRVSLPSQFSSAFS 608
Query: 235 -SRRFSQCLVPFHADSRTSGTISFGDA-----SDVSGEGVVTTPLVSE-----------E 277
R+F+ CL A+S G + FGD DVS + + TPL++ E
Sbjct: 609 FHRKFAICLT---ANSGADGVMFFGDGPYNLNQDVS-KVLTYTPLITNPVSTAPSAFLGE 776
Query: 278 GQTPYLVTLEGISVGDTFVSFNSS 301
Y + ++ I V + V N++
Sbjct: 777 PSVEYFIGVKSIKVSEKNVPLNTT 848
>TC16317 similar to UP|Q8W4C5 (Q8W4C5) Nucellin-like protein, partial (13%)
Length = 1131
Score = 60.5 bits (145), Expect = 5e-10
Identities = 68/270 (25%), Positives = 114/270 (42%), Gaps = 23/270 (8%)
Frame = +2
Query: 186 TNGDELVVGDIIFGCGHSNSGAF----NENDMGVIGLGGGPLSLVSQMGALYGSRRF-SQ 240
TNG + + +FGCG+ +G+ ++ D G++GL +SL Q+ + +
Sbjct: 20 TNGSKTKF-NFVFGCGYDQAGSLLNTLSKTD-GIMGLSRAKVSLPYQLASKGLIKNVVGH 193
Query: 241 CLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTPYLVTLEGISVGDTFVSFNS 300
CL +D+ G + GD V G+ P+ Y + GI+ G+ +S +
Sbjct: 194 CL---SSDAVGGGYMFLGD-DFVPNWGMTWVPMAYTHSSDLYQTEILGINYGNHPLSSDG 361
Query: 301 SEKLSKGNMMIDSGTPATYLPQEFYDRLVEELKVQSSLLPVDNDPDLGTQLCYRSETNLE 360
K+ K ++ DSG+ TY P+E Y LV LK S L + +D D +C++ + +
Sbjct: 362 HSKVRK--VVFDSGSSYTYFPKEAYLDLVASLKEVSGLRLIQDDSDTTFPICWQDDFPIR 535
Query: 361 G--------PILTAHFE------GADVQLMPIQTFIPPKDGVFCFAM---AGTADG-DYI 402
LT F Q+ P I K G C A+ + DG I
Sbjct: 536 SVKDVKDFFKTLTLRFGNKWWILSTFFQIPPEGYLIISKKGNVCLAILDGSNVHDGSSII 715
Query: 403 FGNFAQSNILIGFDLDRKTISFKPTDCTNP 432
G+ + L+ +D + I ++ DC P
Sbjct: 716 LGDVSLRGHLVVYDNVNQRIGWERADCGMP 805
>TC17357 similar to UP|Q9FFC3 (Q9FFC3) Protease-like protein, partial (24%)
Length = 577
Score = 58.5 bits (140), Expect = 2e-09
Identities = 25/68 (36%), Positives = 39/68 (56%), Gaps = 5/68 (7%)
Frame = +1
Query: 83 GDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKS-----PMFEPLSSKTFNP 137
G Y ++ +G+PPV+ Y +DTGSD++W CS C+GC + F+P S T +
Sbjct: 313 GLYFTRVQIGTPPVEFYVQIDTGSDVLWVSCSSCNGCPQTSGLQIQLDFFDPGHSSTSSL 492
Query: 138 IPCDSEQC 145
I C ++C
Sbjct: 493 ISCSDQRC 516
>BF177843
Length = 474
Score = 54.3 bits (129), Expect = 4e-08
Identities = 40/140 (28%), Positives = 59/140 (41%), Gaps = 5/140 (3%)
Frame = +2
Query: 127 FEPLSSKTFNPIPCDSEQCGS-----LFSHSCSPQKLCAYSYSYADSSVTKGVLARETIT 181
F+P S +F+ +PC + C +C + C YS+ YAD ++ +G L E IT
Sbjct: 20 FDPALSSSFSTLPCSNPHCEPGVPDYTIPTTCDRNRHCHYSFFYADGTLAEGNLVTEKIT 199
Query: 182 FSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVSQMGALYGSRRFSQC 241
S L I GC +++S + G++G+ G S SQ +FS C
Sbjct: 200 LSPSQTTPPL-----ILGCANTSS-----DGRGILGMNRGTFSFPSQTKI----AKFSYC 337
Query: 242 LVPFHADSRTSGTISFGDAS 261
VP GT G S
Sbjct: 338 -VPLRQTRPAPGTFLIGQNS 394
>TC16449 similar to UP|Q940R4 (Q940R4) AT4g16560/dl4305c, partial (18%)
Length = 613
Score = 49.7 bits (117), Expect = 9e-07
Identities = 54/181 (29%), Positives = 77/181 (41%), Gaps = 10/181 (5%)
Frame = +3
Query: 1 MANILCFFHLCFLVVL------YLLSCQTPIEAQDAGFSVQLTRQNSPHSPFYKPDNLHR 54
MA+ F LCF++ + LL I + L + S S + HR
Sbjct: 66 MASPSPLFLLCFMLCISHSYQMVLLPLTHSISKSQFNSTHHLLKTTSTRSAARFQHHHHR 245
Query: 55 HKLPSFHQVPKKAFAPNGPFSTRVTSNNGDYLMKLTLG-SPPVDIYGLVDTGSDLVWAQC 113
H+ HQ+ P P S DY + L G S P+ +Y +DTGSDLVW C
Sbjct: 246 HQQ---HQLS----LPLSPGS--------DYTLSLNFGGSQPITLY--MDTGSDLVWFPC 374
Query: 114 SP--CHGCYKQKSPMFEPLSSKTFNPIPCDSEQCGSLFSHSCSPQK-LCAYSYSYADSSV 170
+P C C + K +P + + + C S C + +HS P + LCA S+ DS
Sbjct: 375 APFECMLC-EGKPDTPKPANITKAHTVSCQSPACSA--AHSSMPHRDLCAISHCPLDSIE 545
Query: 171 T 171
T
Sbjct: 546 T 548
>TC12301 weakly similar to UP|CAR5_RHINI (P43232) Rhizopuspepsin 5 precursor
(Aspartate protease) , partial (8%)
Length = 501
Score = 49.7 bits (117), Expect = 9e-07
Identities = 40/162 (24%), Positives = 61/162 (36%), Gaps = 41/162 (25%)
Frame = +2
Query: 80 SNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQC-------------------------- 113
S G+Y +++ +G+P + DTGS+ W
Sbjct: 5 SARGEYFVQVKVGTPGQKFWLAADTGSEFTWFNSVHKTHNKTQTKNHGGGGHKKHKGKLR 184
Query: 114 ---------SPCHGCYKQKSPMFEPLSSKTFNPIPCDSEQC----GSLFS--HSCSPQKL 158
+PC+G +F P S+TF + C S +C LFS + P
Sbjct: 185 KRGKKVRHNNPCNG-------VFCPQRSRTFKTVTCSSRKCKVELSDLFSLTYCPKPSDP 343
Query: 159 CAYSYSYADSSVTKGVLARETITFSSPTNGDELVVGDIIFGC 200
C Y SY D S KG +TIT +NG + + ++ GC
Sbjct: 344 CLYDISYVDGSSAKGFFGSDTITVEL-SNGRKGKLHNLTIGC 466
>AV427004
Length = 428
Score = 48.1 bits (113), Expect = 3e-06
Identities = 50/151 (33%), Positives = 68/151 (44%), Gaps = 4/151 (2%)
Frame = +3
Query: 165 YADSSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSN--SGAFNE-NDMGVIGLGGG 221
YAD + GVL R+ I TNG +V I FGCG+ SG + GVIGLG G
Sbjct: 12 YADHGSSLGVLVRDHIHLHF-TNGS-VVRPKIAFGCGYDQKYSGPITPPSTAGVIGLGNG 185
Query: 222 PLSLVSQMGALYGSRR-FSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQT 280
S+VSQ+ +L R CL ++ G + FGD + G+V TP++ +
Sbjct: 186 RSSIVSQLHSLGLIRNVVGHCL-----SAQGGGFLFFGD-DLIPSSGIVWTPMLPSSMEK 347
Query: 281 PYLVTLEGISVGDTFVSFNSSEKLSKGNMMI 311
Y S G + FN KG +I
Sbjct: 348 HY-------SSGPAELLFNGKPTTVKGLELI 419
>AV411792
Length = 392
Score = 45.8 bits (107), Expect = 1e-05
Identities = 44/130 (33%), Positives = 62/130 (46%), Gaps = 1/130 (0%)
Frame = +3
Query: 168 SSVTKGVLARETITFSSPTNGDELVVGDIIFGCGHSNSGAFNENDMGVIGLGGGPLSLVS 227
SS L R+T+T ++ V FGC +G+ + G++GLG GPLSL++
Sbjct: 6 SSTVAANLVRDTLTLAADR------VPGYTFGCIQKATGS-SVPQQGLLGLGRGPLSLLA 164
Query: 228 QMGALYGSRRFSQCLVPFHADSRTSGTISFGDASDVSGEGVVTTPLVSEEGQTP-YLVTL 286
Q LY S FS CL F A + T G++ G A + TPL+ ++ Y V L
Sbjct: 165 QTQNLYKS-TFSYCLPSFKALNFT-GSLRLGLAQPTR---LKFTPLLKNPRRSSLYYVNL 329
Query: 287 EGISVGDTFV 296
I VG V
Sbjct: 330 VAIKVGRKIV 359
>TC13045 weakly similar to UP|Q9FL43 (Q9FL43) Nucleoid DNA-binding-like
protein, partial (27%)
Length = 446
Score = 44.7 bits (104), Expect = 3e-05
Identities = 22/67 (32%), Positives = 34/67 (49%), Gaps = 1/67 (1%)
Frame = +2
Query: 75 STRVTSNNGDYLMKLTLGSPPVDIYGLVDTGSDLVWAQCSPCHGCYKQKSPM-FEPLSSK 133
S R + + Y+++ +G+PP + +DT +D W C+ C GC SP F P S
Sbjct: 251 SGRQITQSPTYIVRAKIGTPPQTLLLAMDTNNDAAWVPCTGCVGC---SSPTPFSPAKST 421
Query: 134 TFNPIPC 140
TF + C
Sbjct: 422 TFRNLTC 442
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,504,890
Number of Sequences: 28460
Number of extensions: 133733
Number of successful extensions: 846
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 817
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 818
length of query: 432
length of database: 4,897,600
effective HSP length: 93
effective length of query: 339
effective length of database: 2,250,820
effective search space: 763027980
effective search space used: 763027980
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0588b.3


