

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0587.6
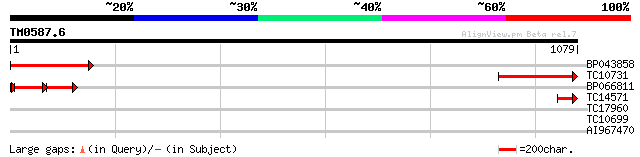
(1079 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP043858 318 2e-87
TC10731 similar to PIR|T51557|T51557 Exportin1 (XPO1) protein - ... 299 2e-81
BP066811 131 2e-58
TC14571 homologue to PIR|T51557|T51557 Exportin1 (XPO1) protein ... 68 9e-12
TC17960 similar to UP|Q8H183 (Q8H183) Beta-ureidopropionase (N-c... 29 4.5
TC10699 similar to UP|MAD2_ARATH (Q9LU93) Mitotic spindle checkp... 29 4.5
AI967470 28 7.7
>BP043858
Length = 546
Score = 318 bits (816), Expect = 2e-87
Identities = 158/158 (100%), Positives = 158/158 (100%)
Frame = +3
Query: 1 MAAEKLRDLSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQN 60
MAAEKLRDLSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQN
Sbjct: 72 MAAEKLRDLSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQN 251
Query: 61 TQNLNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNK 120
TQNLNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNK
Sbjct: 252 TQNLNTKFFALQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNK 431
Query: 121 LNIILVQILKHEWPLRWRSFIPDLVSAAKTSETICENC 158
LNIILVQILKHEWPLRWRSFIPDLVSAAKTSETICENC
Sbjct: 432 LNIILVQILKHEWPLRWRSFIPDLVSAAKTSETICENC 545
>TC10731 similar to PIR|T51557|T51557 Exportin1 (XPO1) protein - Arabidopsis
thaliana (fragment) {Arabidopsis thaliana;}, partial
(14%)
Length = 595
Score = 299 bits (765), Expect = 2e-81
Identities = 149/149 (100%), Positives = 149/149 (100%)
Frame = +3
Query: 931 EIFAVLTDTFHKPGFKLHVLVLQHLFCLAETGALTEPLWDAATNPYPYPSNAAFVLEYTI 990
EIFAVLTDTFHKPGFKLHVLVLQHLFCLAETGALTEPLWDAATNPYPYPSNAAFVLEYTI
Sbjct: 3 EIFAVLTDTFHKPGFKLHVLVLQHLFCLAETGALTEPLWDAATNPYPYPSNAAFVLEYTI 182
Query: 991 KLLSTSFPNMTAAEVTQFVNGLFESTNDLSTFKNHIRDFLVQSKEFSAQDNKDLYAEEAA 1050
KLLSTSFPNMTAAEVTQFVNGLFESTNDLSTFKNHIRDFLVQSKEFSAQDNKDLYAEEAA
Sbjct: 183 KLLSTSFPNMTAAEVTQFVNGLFESTNDLSTFKNHIRDFLVQSKEFSAQDNKDLYAEEAA 362
Query: 1051 AQRERERQRMLSVPGLIAPSELQDEMLDS 1079
AQRERERQRMLSVPGLIAPSELQDEMLDS
Sbjct: 363 AQRERERQRMLSVPGLIAPSELQDEMLDS 449
>BP066811
Length = 460
Score = 131 bits (329), Expect(3) = 2e-58
Identities = 64/64 (100%), Positives = 64/64 (100%)
Frame = +3
Query: 9 LSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKF 68
LSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKF
Sbjct: 78 LSQPIDVPLLDATVAAFYGTGSKEQRTAADQILRDLQNNPDMWLQVMHILQNTQNLNTKF 257
Query: 69 FALQ 72
FALQ
Sbjct: 258 FALQ 269
Score = 113 bits (282), Expect(3) = 2e-58
Identities = 56/58 (96%), Positives = 57/58 (97%)
Frame = +2
Query: 71 LQVLEGVIKYRWNALPLEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQI 128
+QVLEGVIKYRWNALPLEQRDGMKNFISDI VQLSGNEASFRMERLYVNKLNIILVQI
Sbjct: 287 IQVLEGVIKYRWNALPLEQRDGMKNFISDIXVQLSGNEASFRMERLYVNKLNIILVQI 460
Score = 21.2 bits (43), Expect(3) = 2e-58
Identities = 9/12 (75%), Positives = 9/12 (75%)
Frame = +1
Query: 1 MAAEKLRDLSQP 12
MAAEKLRD P
Sbjct: 55 MAAEKLRDCPNP 90
>TC14571 homologue to PIR|T51557|T51557 Exportin1 (XPO1) protein - Arabidopsis
thaliana (fragment) {Arabidopsis thaliana;}, partial (3%)
Length = 677
Score = 67.8 bits (164), Expect = 9e-12
Identities = 32/37 (86%), Positives = 36/37 (96%)
Frame = +2
Query: 1043 DLYAEEAAAQRERERQRMLSVPGLIAPSELQDEMLDS 1079
DLYAEEAAAQRERER RML++PGLIAP+ELQDEM+DS
Sbjct: 2 DLYAEEAAAQRERERPRMLAIPGLIAPNELQDEMVDS 112
>TC17960 similar to UP|Q8H183 (Q8H183) Beta-ureidopropionase
(N-carbamyl-beta-alanine amidohydrolase) (At5g64370) ,
partial (29%)
Length = 455
Score = 28.9 bits (63), Expect = 4.5
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Frame = -3
Query: 671 KWLEIIGQARQNVDFL--KDHDVI--RTVLNILQTNTSVASSLGTYF 713
KW I ++Q VDFL ++I + V ++ T+ SVA LG +F
Sbjct: 207 KWFSTIKPSKQAVDFLIVMGFEIILEKMVKRVVATDRSVAGGLGLFF 67
>TC10699 similar to UP|MAD2_ARATH (Q9LU93) Mitotic spindle checkpoint
protein MAD2, partial (95%)
Length = 975
Score = 28.9 bits (63), Expect = 4.5
Identities = 24/104 (23%), Positives = 49/104 (47%), Gaps = 9/104 (8%)
Frame = +2
Query: 75 EGVIKYRWNALP--LEQRDGMKNFISDIIVQLSGNEASFRMERLYVNKLNIILVQILKHE 132
E +K + LP L + +G+K+FIS++ QLS S +++R+ + ++ ++L+
Sbjct: 140 ESFVKVKKYGLPMLLTEDEGVKSFISNLTAQLSEWLESGKLQRVVLVIMSKATGEVLE-- 313
Query: 133 WPLRWR-------SFIPDLVSAAKTSETICENCMAILKLLSEEV 169
RW + VS K+ + I AI+ ++ +
Sbjct: 314 ---RWNFSIETDSEVVEKSVSREKSDKEIMREIQAIMSQIASSI 436
>AI967470
Length = 356
Score = 28.1 bits (61), Expect = 7.7
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Frame = +3
Query: 146 SAAKTSETICE---NCMAILKLLSEEVFDFSRGEMTQQK 181
S AKTS+T C NC IL LSE + ++ +T Q+
Sbjct: 240 SEAKTSKTDCSLSSNCGKILASLSEALIHYATSSITPQQ 356
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,729,600
Number of Sequences: 28460
Number of extensions: 222486
Number of successful extensions: 1149
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 1143
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1149
length of query: 1079
length of database: 4,897,600
effective HSP length: 100
effective length of query: 979
effective length of database: 2,051,600
effective search space: 2008516400
effective search space used: 2008516400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0587.6


