

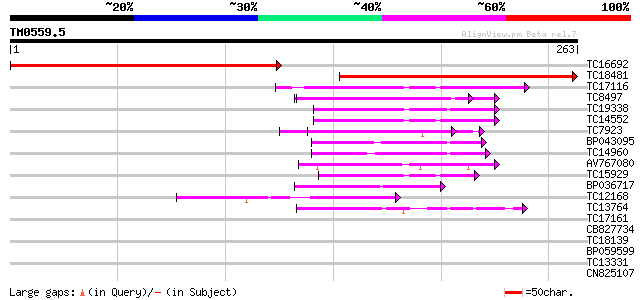
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0559.5
(263 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16692 weakly similar to UP|O23166 (O23166) Thiol-disulfide int... 254 1e-68
TC18481 similar to UP|O23166 (O23166) Thiol-disulfide interchang... 222 4e-59
TC17116 weakly similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-t... 54 2e-08
TC8497 similar to UP|PDA6_MEDSA (P38661) Probable protein disulf... 53 6e-08
TC19338 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m ... 52 8e-08
TC14552 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, ... 51 2e-07
TC7923 similar to UP|PDI_MEDSA (P29828) Protein disulfide isomer... 49 7e-07
BP043095 47 3e-06
TC14960 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%) 47 3e-06
AV767080 44 2e-05
TC15929 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, ... 44 3e-05
BP036717 43 6e-05
TC12168 similar to UP|Q8L7S9 (Q8L7S9) At1g43560/T10P12_4, partia... 43 6e-05
TC13764 similar to UP|Q9C9Y6 (Q9C9Y6) Thioredoxin-like protein, ... 42 8e-05
TC17161 homologue to UP|THIF_PEA (P29450) Thioredoxin F-type, ch... 35 0.017
CB827734 33 0.038
TC18139 32 0.15
BP059599 31 0.19
TC13331 similar to UP|Q8LE11 (Q8LE11) Ripening-related protein-l... 30 0.42
CN825107 27 4.7
>TC16692 weakly similar to UP|O23166 (O23166) Thiol-disulfide interchange
like protein (Thioredoxin-like protein)
(AT4g37200/C7A10_160), partial (16%)
Length = 415
Score = 254 bits (648), Expect = 1e-68
Identities = 126/126 (100%), Positives = 126/126 (100%)
Frame = +1
Query: 1 MSRLSSNPIGLHRFRPCLRTPQFTVNPRHLHFNTRKFHTLACQTNPNLDEKDASATSQEI 60
MSRLSSNPIGLHRFRPCLRTPQFTVNPRHLHFNTRKFHTLACQTNPNLDEKDASATSQEI
Sbjct: 37 MSRLSSNPIGLHRFRPCLRTPQFTVNPRHLHFNTRKFHTLACQTNPNLDEKDASATSQEI 216
Query: 61 KIVVEPSTVNGESETCKPTSSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRLDFG 120
KIVVEPSTVNGESETCKPTSSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRLDFG
Sbjct: 217 KIVVEPSTVNGESETCKPTSSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRLDFG 396
Query: 121 VSLKDL 126
VSLKDL
Sbjct: 397 VSLKDL 414
>TC18481 similar to UP|O23166 (O23166) Thiol-disulfide interchange like
protein (Thioredoxin-like protein)
(AT4g37200/C7A10_160), partial (42%)
Length = 602
Score = 222 bits (566), Expect = 4e-59
Identities = 108/110 (98%), Positives = 109/110 (98%)
Frame = +2
Query: 154 VCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNV 213
VCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNV
Sbjct: 2 VCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFLDKEGNEEGNV 181
Query: 214 VGRLPRQLLLENVDALARGEASVPHARVVGKYSSAEARKVHQVVDPRSHG 263
VGRLPRQLLLENVDALARGEASVPHARVVG+YSSAEARKVH VVDPRSHG
Sbjct: 182 VGRLPRQLLLENVDALARGEASVPHARVVGQYSSAEARKVHPVVDPRSHG 331
>TC17116 weakly similar to UP|THM3_ARATH (Q9SEU7) Thioredoxin M-type 3,
chloroplast precursor (TRX-M3), partial (55%)
Length = 798
Score = 54.3 bits (129), Expect = 2e-08
Identities = 31/118 (26%), Positives = 61/118 (51%)
Frame = +3
Query: 124 KDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNT 183
KDL N++ L + P +VEFYA WC CR + + I ++Y R+N ++N D
Sbjct: 183 KDLWENSI-----LKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 347
Query: 184 NWEQELDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHARV 241
Q D++ ++ +P L K+G + V+G +P++ + ++ + + +P ++
Sbjct: 348 --MQIADDYEIKAVP-VVMLFKDGQKCDAVIGTMPKEFYVNAIERVLKP*T*IPADKI 512
>TC8497 similar to UP|PDA6_MEDSA (P38661) Probable protein disulfide
isomerase A6 precursor (P5) , partial (95%)
Length = 1533
Score = 52.8 bits (125), Expect = 6e-08
Identities = 28/82 (34%), Positives = 40/82 (48%)
Frame = +2
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFG 193
E L K +VEFYA WC C+ LAP K+ +K + V+ N+D + +++
Sbjct: 581 EVVLDETKDVLVEFYAPWCGHCKSLAPTYEKVAAAFKLDGDVVIANLDADKYRDLAEKYE 760
Query: 194 VEGIPHFAFLDKEGNEEGNVVG 215
V G P F K GN+ G G
Sbjct: 761 VSGFPTLKFFPK-GNKAGEEYG 823
Score = 51.2 bits (121), Expect = 2e-07
Identities = 27/95 (28%), Positives = 45/95 (46%)
Frame = +2
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
+E+ + K +VEFYA WC C++LAP+ K+ +K + ++ VD + ++
Sbjct: 221 FEKEVGQDKGALVEFYAPWCGHCKKLAPEYEKLGGSFKKAKSVLIAKVDCDEHKSVCSKY 400
Query: 193 GVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVD 227
GV G P + K E G + L E V+
Sbjct: 401 GVSGYPTLQWFPKGSLEPKKYEGPRTAEALAEFVN 505
>TC19338 similar to GB|AAC49358.1|1388088|PSU35831 thioredoxin m {Pisum
sativum;} , partial (65%)
Length = 476
Score = 52.4 bits (124), Expect = 8e-08
Identities = 24/86 (27%), Positives = 49/86 (56%)
Frame = +2
Query: 142 PTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFA 201
P +V+F+A WC CR +AP + ++ ++Y ++ LN D++ ++G+ IP
Sbjct: 86 PVLVDFWAPWCGPCRMIAPLIDELAKEYAGKIACYKLNTDDS--PNIATQYGIRSIPTVL 259
Query: 202 FLDKEGNEEGNVVGRLPRQLLLENVD 227
F K G ++ +V+G +P+ L +++
Sbjct: 260 FF-KNGEKKESVIGAVPKSTLTASLE 334
>TC14552 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (55%)
Length = 1016
Score = 51.2 bits (121), Expect = 2e-07
Identities = 23/86 (26%), Positives = 46/86 (52%)
Frame = +1
Query: 142 PTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFA 201
P +VEF+A WC CR + P + ++ +QY ++ LN D + +G+ IP
Sbjct: 355 PVLVEFWAPWCGPCRMIHPIIDELAKQYAGKLKCYKLNTDES--PSTATRYGIRSIP-TV 525
Query: 202 FLDKEGNEEGNVVGRLPRQLLLENVD 227
+ ++G ++ V+G +P+ L +++
Sbjct: 526 MIFRDGEKKDTVIGAVPKSTLTSSIE 603
>TC7923 similar to UP|PDI_MEDSA (P29828) Protein disulfide isomerase
precursor (PDI) , partial (89%)
Length = 1861
Score = 49.3 bits (116), Expect = 7e-07
Identities = 22/69 (31%), Positives = 37/69 (52%)
Frame = +2
Query: 139 NGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIP 198
+GK ++EFYA WC C+ LAP + ++ Y++ + V+ D T + + F V+G P
Sbjct: 1226 SGKNVLLEFYAPWCGHCKSLAPILDEVAVSYQNDADIVIAKFDATANDVPTESFEVQGYP 1405
Query: 199 HFAFLDKEG 207
F+ G
Sbjct: 1406 TLYFISSSG 1432
Score = 39.3 bits (90), Expect = 7e-04
Identities = 28/98 (28%), Positives = 41/98 (41%), Gaps = 3/98 (3%)
Frame = +2
Query: 126 LTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNW 185
LT + + + +S VVEFYA WC C+ LAP+ K ++L + N
Sbjct: 152 LTLDNSNFSEIVSKHDFIVVEFYAPWCGHCKNLAPEYEKAAAVLSSHDPPIVLAKVDANE 331
Query: 186 EQELD---EFGVEGIPHFAFLDKEGNEEGNVVGRLPRQ 220
E D ++ V G P L G + G PR+
Sbjct: 332 ENNKDLASQYEVRGFPTIKILRNGGKDSQEYKG--PRE 439
>BP043095
Length = 396
Score = 47.0 bits (110), Expect = 3e-06
Identities = 28/81 (34%), Positives = 43/81 (52%)
Frame = +1
Query: 141 KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHF 200
K VV+F A WC CR +AP + ++ ++Y N + L VD + ++ VE +P F
Sbjct: 145 KLIVVDFTASWCGPCRFIAPYLGELAKKY---TNVIFLKVDVDELKSVAQDWAVEAMPTF 315
Query: 201 AFLDKEGNEEGNVVGRLPRQL 221
F+ KEG+ VVG +L
Sbjct: 316 MFV-KEGSIVDKVVGAKKEEL 375
>TC14960 similar to UP|Q8GUR9 (Q8GUR9) Thioredoxin h, partial (98%)
Length = 688
Score = 47.0 bits (110), Expect = 3e-06
Identities = 32/83 (38%), Positives = 42/83 (50%)
Frame = +1
Query: 141 KPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHF 200
K VV+F A WC CR +AP + +I K + V L VD + +E+ VE +P F
Sbjct: 163 KLIVVDFTASWCGPCRFIAPILAEIA---KKLPHVVFLKVDVDELKTVAEEWNVEAMPTF 333
Query: 201 AFLDKEGNEEGNVVGRLPRQLLL 223
FL KEG VVG +L L
Sbjct: 334 LFL-KEGKAVDKVVGAKKEELQL 399
>AV767080
Length = 559
Score = 44.3 bits (103), Expect = 2e-05
Identities = 27/98 (27%), Positives = 50/98 (50%), Gaps = 5/98 (5%)
Frame = -1
Query: 135 QALSNGK---PTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQEL-D 190
Q L GK P +++FYA WC C +A ++ + +Y+ V V ++ D+ E E
Sbjct: 550 QDLVKGKXDVPLIIDFYATWCGPCILMAQELEMLAVEYEQNVTIVKVDTDD---EYEFAR 380
Query: 191 EFGVEGIPHFAFLDKEGNEEG-NVVGRLPRQLLLENVD 227
+ V G+P F+ + N++ G +P Q++ + +D
Sbjct: 379 DMQVRGLPTVFFISPDPNKDAIRTEGLIPIQMMRDIID 266
>TC15929 similar to UP|THM4_ARATH (Q9SEU6) Thioredoxin M-type 4, chloroplast
precursor (TRX-M4), partial (49%)
Length = 1406
Score = 43.9 bits (102), Expect = 3e-05
Identities = 21/75 (28%), Positives = 40/75 (53%)
Frame = +1
Query: 144 VVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEFGVEGIPHFAFL 203
+VEF+A WC CR + P + ++ ++Y ++ LN D + +G+ IP +
Sbjct: 364 LVEFWAPWCGPCRMIHPVIDELAKEYAGKLKCYKLNTDES--PSTATRYGIRSIP-TVII 534
Query: 204 DKEGNEEGNVVGRLP 218
K+G ++ V+G +P
Sbjct: 535 FKDGEKKDAVIGSVP 579
>BP036717
Length = 477
Score = 42.7 bits (99), Expect = 6e-05
Identities = 22/70 (31%), Positives = 34/70 (48%)
Frame = +1
Query: 133 YEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVDNTNWEQELDEF 192
+ + N + +VEFYA WC C+ LAP+ + K+ V+ VD T E+
Sbjct: 175 FTTVVENNRFVMVEFYAPWCGHCQSLAPEYAAAATELKND-GVVLAKVDATQENDLAHEY 351
Query: 193 GVEGIPHFAF 202
V+G P +F
Sbjct: 352 DVQGFPTVSF 381
>TC12168 similar to UP|Q8L7S9 (Q8L7S9) At1g43560/T10P12_4, partial (50%)
Length = 408
Score = 42.7 bits (99), Expect = 6e-05
Identities = 30/107 (28%), Positives = 51/107 (47%), Gaps = 3/107 (2%)
Frame = +1
Query: 78 PTSSTADASGLPQLPTKDINRKVAIASTLAA---LGLFLATRLDFGVSLKDLTANALPYE 134
P+SST +S P L R + I ++ A L + A + F S D+ AN+
Sbjct: 94 PSSSTRLSSTTPSLHFPSRTRVLRIGTSRAPPRFLPVVQAKKQTFS-SFDDMLANS---- 258
Query: 135 QALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVD 181
KP V+FYA WC C+ + P + ++ + KD++ V ++ +
Sbjct: 259 -----DKPVFVDFYATWCGPCQFMVPILDEVSTRLKDKIQVVKIDTE 384
>TC13764 similar to UP|Q9C9Y6 (Q9C9Y6) Thioredoxin-like protein, partial
(73%)
Length = 585
Score = 42.4 bits (98), Expect = 8e-05
Identities = 33/112 (29%), Positives = 59/112 (52%), Gaps = 5/112 (4%)
Frame = +3
Query: 134 EQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNFVMLNVD-----NTNWEQE 188
E+A +GK + F A WC C+ +AP ++ ++Y + F++++VD +T+W+
Sbjct: 12 EEAKRDGKIVIANFSATWCGPCKIIAPYYCELSEKYTSMM-FLLVDVDELTDFSTSWD-- 182
Query: 189 LDEFGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALARGEASVPHAR 240
++ P F FL K+G + +VG R L + V A+A SVP ++
Sbjct: 183 -----IKATPTFFFL-KDGQQLDQLVG-ANRPELEKKVVAIA---DSVPESK 308
>TC17161 homologue to UP|THIF_PEA (P29450) Thioredoxin F-type, chloroplast
precursor (TRX-F), partial (72%)
Length = 850
Score = 34.7 bits (78), Expect = 0.017
Identities = 39/152 (25%), Positives = 67/152 (43%), Gaps = 5/152 (3%)
Frame = +2
Query: 57 SQEIKIVVEPSTV---NGESETCKPTSSTADASGLPQLPTKDINRK--VAIASTLAALGL 111
S + K V PST+ N S SS++ +S + + R VA+ S+L G
Sbjct: 59 SPKCKWTVPPSTIVFDNASSSKLPLGSSSSSSSASSVYRSVSLRRSGSVAVRSSLETAGP 238
Query: 112 FLATRLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKD 171
+ V+ KD P A + K V++ + WC C+ +AP ++ ++ D
Sbjct: 239 TVTVGKVTEVN-KD---TFWPIVNAAGD-KTVVLDMFTQWCGPCKVIAPKYQELAEKNLD 403
Query: 172 RVNFVMLNVDNTNWEQELDEFGVEGIPHFAFL 203
V F+ L+ + N + E G++ +P F L
Sbjct: 404 -VVFLKLDCNQDN-KPLAKELGIKVVPTFKIL 493
>CB827734
Length = 556
Score = 33.5 bits (75), Expect = 0.038
Identities = 23/79 (29%), Positives = 34/79 (42%), Gaps = 1/79 (1%)
Frame = +1
Query: 119 FGVSLKDLTANALPYEQALSNGKPTV-VEFYADWCEVCRELAPDVYKIEQQYKDRVNFVM 177
+G S + N ++ + N K V VEF+A WC C+ L P K K V
Sbjct: 118 YGASSPVVQLNPSNFKSKVLNSKGVVLVEFFAPWCGHCKALTPIWEKAATVLKGVVTVAA 297
Query: 178 LNVDNTNWEQELDEFGVEG 196
L+ D + E+G+ G
Sbjct: 298 LDAD--AHQALAQEYGIRG 348
>TC18139
Length = 429
Score = 31.6 bits (70), Expect = 0.15
Identities = 30/127 (23%), Positives = 54/127 (41%), Gaps = 5/127 (3%)
Frame = +1
Query: 116 RLDFGVSLKDLTANALPYEQALSNGKPTVVEFYADWCEVCRELAPDVYKIEQQYKDRVNF 175
R G S++D+ + A ++ + G + F+A WCE + + + + +F
Sbjct: 19 RSRMGGSVRDVKSKA-ELDEVVHGGAAVALHFWASWCEASKHMDKIFSHLSTDF-PHAHF 192
Query: 176 VMLNVDNTNWEQELDE-FGVEGIPHFAFLDKEGNEEGNVVGRLPRQLLLENVDALAR--- 231
+ + + + E+ E + V +P F F K+G G P L V +AR
Sbjct: 193 LRVEAEE---QPEISEAYSVSAVPFFVFC-KDGKTVDTSEGADPSS-LANKVAKVARSVN 357
Query: 232 -GEASVP 237
GEA+ P
Sbjct: 358 PGEAASP 378
>BP059599
Length = 367
Score = 31.2 bits (69), Expect = 0.19
Identities = 22/88 (25%), Positives = 35/88 (39%)
Frame = +3
Query: 1 MSRLSSNPIGLHRFRPCLRTPQFTVNPRHLHFNTRKFHTLACQTNPNLDEKDASATSQEI 60
+S S+NP +PCL P F + +H++ + H LA P S +
Sbjct: 6 LSSSSNNPEKSKXKKPCLEPPLFLIF-KHIYHHNPGTHKLAPTMEPTNLSS*TQHPSSSL 182
Query: 61 KIVVEPSTVNGESETCKPTSSTADASGL 88
V P +N + K S + SG+
Sbjct: 183CTTVTPELLNQSPHSIKSPRSKPNRSGV 266
>TC13331 similar to UP|Q8LE11 (Q8LE11) Ripening-related protein-like,
partial (7%)
Length = 826
Score = 30.0 bits (66), Expect = 0.42
Identities = 19/56 (33%), Positives = 28/56 (49%)
Frame = +1
Query: 67 STVNGESETCKPTSSTADASGLPQLPTKDINRKVAIASTLAALGLFLATRLDFGVS 122
STV +C + +DA +L D+NR+V + L + L+L TRL G S
Sbjct: 511 STVMSNGGSCVRSFRNSDADAKLKL---DMNRRVTTVTMLTSNALYLITRLSEGTS 669
>CN825107
Length = 254
Score = 26.6 bits (57), Expect = 4.7
Identities = 15/45 (33%), Positives = 22/45 (48%)
Frame = -2
Query: 54 SATSQEIKIVVEPSTVNGESETCKPTSSTADASGLPQLPTKDINR 98
S++S PST SETC TS ++ S + LP K+ +
Sbjct: 193 SSSSSSPSSSFSPSTTYFSSETCN-TSPSSSTSSIESLPLKNFKK 62
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,131,775
Number of Sequences: 28460
Number of extensions: 51024
Number of successful extensions: 268
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 265
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 268
length of query: 263
length of database: 4,897,600
effective HSP length: 88
effective length of query: 175
effective length of database: 2,393,120
effective search space: 418796000
effective search space used: 418796000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0559.5


