

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0545.2
(653 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
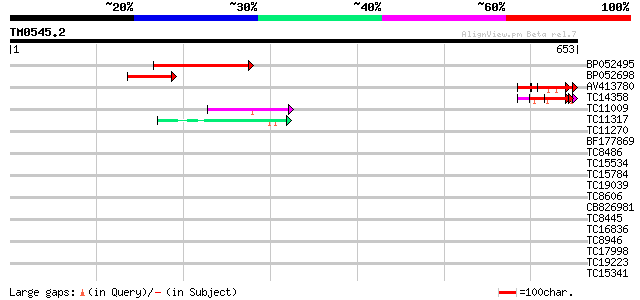
Score E
Sequences producing significant alignments: (bits) Value
BP052495 181 4e-46
BP052698 88 4e-18
AV413780 72 2e-13
TC14358 60 8e-10
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 45 5e-05
TC11317 similar to UP|Q40363 (Q40363) NuM1 protein, partial (12%) 44 8e-05
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 40 0.001
BF177869 40 0.001
TC8486 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Ara... 40 0.002
TC15534 weakly similar to GB|AAO63420.1|28950993|BT005356 At5g28... 39 0.002
TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precurso... 39 0.003
TC19039 similar to UP|Q8L7W5 (Q8L7W5) AT4g12110/F16J13_180, part... 39 0.003
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 38 0.004
CB826981 37 0.008
TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial ... 37 0.013
TC16836 similar to GB|AAB68983.1|500656|YSCH9666 Yhr143wp {Sacch... 37 0.013
TC8946 homologue to GB|AAM66972.1|21617922|AY088650 pyridoxine b... 36 0.017
TC17998 homologue to UP|Q8UKS4 (Q8UKS4) IS21 family transposase,... 36 0.022
TC19223 similar to UP|Q940C3 (Q940C3) AT3g27530/MMJ24_7, partial... 35 0.050
TC15341 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pro... 33 0.19
>BP052495
Length = 524
Score = 181 bits (458), Expect = 4e-46
Identities = 96/116 (82%), Positives = 104/116 (88%), Gaps = 1/116 (0%)
Frame = -3
Query: 166 VALADKLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVD 225
V L D L + ++RKRGATPTTSASQKRAKGAG + GNPSPAKSASQREE+QQGG+QGVD
Sbjct: 429 VYLCDHLLLFQSRKRGATPTTSASQKRAKGAGEPSAGNPSPAKSASQREEVQQGGEQGVD 250
Query: 226 QQASSP-LQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPK 280
+QAS+P LQ TT IPVEMAQAKKTSGKR SRKSGSSSSHHSKSSTSSSPLKASGPK
Sbjct: 249 EQASNPLLQGTTPIPVEMAQAKKTSGKRYSRKSGSSSSHHSKSSTSSSPLKASGPK 82
Score = 38.5 bits (88), Expect = 0.003
Identities = 17/18 (94%), Positives = 17/18 (94%)
Frame = -1
Query: 159 EDDDDDDVALADKLKIPK 176
EDDDDDDVAL DKLKIPK
Sbjct: 524 EDDDDDDVALPDKLKIPK 471
>BP052698
Length = 499
Score = 88.2 bits (217), Expect = 4e-18
Identities = 44/57 (77%), Positives = 50/57 (87%)
Frame = +1
Query: 136 QAEPIPRPTPQASPRVASATVEMEDDDDDDVALADKLKIPKTRKRGATPTTSASQKR 192
+AEPIPRPTPQASPRV SATVE+E DDDDDV+LADKLK+ K++KRGATP S S KR
Sbjct: 331 KAEPIPRPTPQASPRVTSATVEIE-DDDDDVSLADKLKVXKSQKRGATPNNSXSPKR 498
>AV413780
Length = 290
Score = 72.4 bits (176), Expect = 2e-13
Identities = 37/45 (82%), Positives = 40/45 (88%)
Frame = +1
Query: 609 EDIFVRSSEDQKIRGSEDLKIRGSEDQKIRGSEDLKIRGSEDQRI 653
ED+ +R SEDQKIR SEDL+IRGSEDQKI GSEDLKIR SEDQRI
Sbjct: 124 EDLKIRRSEDQKIRRSEDLRIRGSEDQKI*GSEDLKIRRSEDQRI 258
Score = 68.9 bits (167), Expect = 2e-12
Identities = 43/76 (56%), Positives = 47/76 (61%), Gaps = 24/76 (31%)
Frame = +2
Query: 602 VVLILTPEDIFVRSSEDQ------------------------KIRGSEDLKIRGSEDQKI 637
+VLILTPEDIFVR SEDQ KI GSEDLKIR SEDQ+I
Sbjct: 47 LVLILTPEDIFVRRSEDQRI*RSEDQRI*RSEDLKIRRSEDQKI*GSEDLKIRRSEDQRI 226
Query: 638 RGSEDLKIRGSEDQRI 653
SEDL+IRGS+DQRI
Sbjct: 227 *RSEDLRIRGSDDQRI 274
Score = 63.9 bits (154), Expect = 8e-11
Identities = 33/45 (73%), Positives = 38/45 (84%)
Frame = +3
Query: 609 EDIFVRSSEDQKIRGSEDLKIRGSEDQKIRGSEDLKIRGSEDQRI 653
ED +R S+DQ+I SEDL+IRGSEDQKI GSEDL IRGSEDQ+I
Sbjct: 156 EDQKIRRSKDQRI*RSEDLRIRGSEDQKI*GSEDLMIRGSEDQKI 290
Score = 55.1 bits (131), Expect = 4e-08
Identities = 35/61 (57%), Positives = 38/61 (61%), Gaps = 8/61 (13%)
Frame = +1
Query: 601 HVVLILTPEDIFVRSSEDQKIRGSEDLK--------IRGSEDQKIRGSEDLKIRGSEDQR 652
H +LI D R QKI GSEDLK IR SEDQKIR SEDL+IRGSEDQ+
Sbjct: 28 HHLLIFLGSDSDTRRYIRQKI*GSEDLKIRRSEDLKIRRSEDQKIRRSEDLRIRGSEDQK 207
Query: 653 I 653
I
Sbjct: 208 I 210
Score = 54.3 bits (129), Expect = 6e-08
Identities = 30/62 (48%), Positives = 42/62 (67%)
Frame = +1
Query: 585 QSIETKKAQVVRSRFGHVVLILTPEDIFVRSSEDQKIRGSEDLKIRGSEDQKIRGSEDLK 644
+ ++ ++++ ++ R I ED+ +R SEDQKI GSEDLKIR SEDQ+I SED K
Sbjct: 100 EDLKIRRSEDLKIRRSEDQKIRRSEDLRIRGSEDQKI*GSEDLKIRRSEDQRI**SEDQK 279
Query: 645 IR 646
IR
Sbjct: 280 IR 285
Score = 53.9 bits (128), Expect = 8e-08
Identities = 30/45 (66%), Positives = 34/45 (74%)
Frame = +1
Query: 609 EDIFVRSSEDQKIRGSEDLKIRGSEDQKIRGSEDLKIRGSEDQRI 653
ED +R SED +IRGSED KI GSED KIR SED +I SEDQ+I
Sbjct: 148 EDQKIRRSEDLRIRGSEDQKI*GSEDLKIRRSEDQRI**SEDQKI 282
Score = 29.3 bits (64), Expect = 2.1
Identities = 19/53 (35%), Positives = 29/53 (53%)
Frame = +1
Query: 578 DKAIAVEQSIETKKAQVVRSRFGHVVLILTPEDIFVRSSEDQKIRGSEDLKIR 630
D I + + ++++ +R R I ED+ +R SEDQ+I SED KIR
Sbjct: 127 DLKIRRSEDQKIRRSEDLRIRGSEDQKI*GSEDLKIRRSEDQRI**SEDQKIR 285
>TC14358
Length = 1084
Score = 60.5 bits (145), Expect = 8e-10
Identities = 33/47 (70%), Positives = 34/47 (72%)
Frame = +1
Query: 599 FGHVVLILTPEDIFVRSSEDQKIRGSEDLKIRGSEDQKIRGSEDLKI 645
F VLILTPEDIF+R SEDQK SEDLK R SED K R SED KI
Sbjct: 175 FDKQVLILTPEDIFIRRSEDQKF*RSEDLKTRRSEDLKTRRSEDQKI 315
Score = 48.9 bits (115), Expect = 3e-06
Identities = 39/96 (40%), Positives = 49/96 (50%), Gaps = 28/96 (29%)
Frame = +2
Query: 586 SIETKKAQVVRSRFGHVV----LILTPEDIFVRSSED----------------QKIRGSE 625
S+ T VRSRFGHVV L++T +++ + + QKI SE
Sbjct: 44 SVITSIGASVRSRFGHVVQLYVLMITSIYLWMNNYDTLTFVLSVLTNRF*F*HQKIYSSE 223
Query: 626 DLKIRGSEDQKIRGSEDLKI--------RGSEDQRI 653
DLKIR SEDQKI EDLKI R SEDQ++
Sbjct: 224 DLKIRSSEDQKI*RPEDLKI*RPEDLRIRRSEDQKL 331
Score = 43.1 bits (100), Expect = 1e-04
Identities = 25/34 (73%), Positives = 25/34 (73%)
Frame = +3
Query: 616 SEDQKIRGSEDLKIRGSEDQKIRGSEDLKIRGSE 649
SE KIR SED KI SEDQKI GSEDLK R SE
Sbjct: 234 SEVLKIRRSEDQKI*RSEDQKI*GSEDLKTRSSE 335
Score = 37.7 bits (86), Expect = 0.006
Identities = 21/33 (63%), Positives = 23/33 (69%)
Frame = +3
Query: 609 EDIFVRSSEDQKIRGSEDLKIRGSEDQKIRGSE 641
E + +R SEDQKI SED KI GSED K R SE
Sbjct: 237 EVLKIRRSEDQKI*RSEDQKI*GSEDLKTRSSE 335
Score = 35.8 bits (81), Expect = 0.022
Identities = 27/54 (50%), Positives = 30/54 (55%), Gaps = 6/54 (11%)
Frame = +3
Query: 602 VVLILTPEDIFVRSSEDQKIRGSEDLKIRGSEDQKIRGSEDLKI------RGSE 649
V+ I ED + SEDQKI GSEDLK R SE G E LK+ RGSE
Sbjct: 240 VLKIRRSEDQKI*RSEDQKI*GSEDLKTRSSE-----GPEALKVQKL*RTRGSE 386
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 44.7 bits (104), Expect = 5e-05
Identities = 37/103 (35%), Positives = 52/103 (49%), Gaps = 5/103 (4%)
Frame = -2
Query: 229 SSPLQDTTSIPVEMAQAKKTSGKRSSRKS--GSSSSHHSKSSTSSSPLKASG---PKRVG 283
SSP + S ++ + + + R +R S SSSS S SS+SSS +SG P R
Sbjct: 327 SSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPP 148
Query: 284 MSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESDPV 326
+ + ++ SS PP SSSSS SSSS S+ S P+
Sbjct: 147 STPPSSSSSSSSSSSSPPPGASSSSSSSPSSSSPSSSSSSSPL 19
Score = 32.0 bits (71), Expect = 0.32
Identities = 25/62 (40%), Positives = 31/62 (49%), Gaps = 1/62 (1%)
Frame = -2
Query: 265 SKSSTS-SSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
S SS S SSP +SG K + S + + P + SSSSS SSSSS S+ S
Sbjct: 348 SPSSLSLSSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGS 169
Query: 324 DP 325
P
Sbjct: 168 PP 163
>TC11317 similar to UP|Q40363 (Q40363) NuM1 protein, partial (12%)
Length = 547
Score = 43.9 bits (102), Expect = 8e-05
Identities = 46/172 (26%), Positives = 70/172 (39%), Gaps = 18/172 (10%)
Frame = +1
Query: 171 KLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASS 230
++K+PK+ K+ AT +A+ A P AK A + E + V++Q S+
Sbjct: 73 RVKMPKSSKKSATKVEAAAPVAAA---------PKSAKKAKRAAEDE------VEKQVSA 207
Query: 231 PLQDTTSIPVEMAQAKKTSG-KRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAV 289
Q + + + K K+ S SSS K + +PLK S +K A
Sbjct: 208 KKQKIAEVAAKQKKETKLQKVKKESSSDDDSSSEDEKPAPKVAPLKTSVKNGTTPAKKAK 387
Query: 290 PRATWFSS--------------KPPPIAL---SSSSSDLESSSSSLDSTESD 324
P + SS KP + + SSSD +SSS S+ESD
Sbjct: 388 PAPSSSSSDEDSSDEEEEVIAKKPTKVVVPKKEESSSDEDSSSDEDSSSESD 543
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 40.0 bits (92), Expect = 0.001
Identities = 40/120 (33%), Positives = 54/120 (44%), Gaps = 2/120 (1%)
Frame = -3
Query: 222 QGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSG--SSSSHHSKSSTSSSPLKASGP 279
+GV + SPL+ +T +P +S SS S SSS+ S SS+SSSP +S
Sbjct: 604 EGVPEPEWSPLE-STFLPFTSTTTTPSSSSSSSSSSFPCSSSTTTSSSSSSSSPFSSSSS 428
Query: 280 KRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSSLDSTESDPVWDKLTFYFNSKPI 339
S +PR + SSSS SSSSS ST L+F+F P+
Sbjct: 427 SSCACSTRDLPRV--------GCGVCSSSS---SSSSSSSSTPWFSFSTSLSFFFLPPPL 281
>BF177869
Length = 509
Score = 40.0 bits (92), Expect = 0.001
Identities = 23/68 (33%), Positives = 38/68 (55%)
Frame = +2
Query: 204 PSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSH 263
P+P + + ++++ GD+ VD P+ + PVE+ + K + +S SGSSSS
Sbjct: 95 PAPVEVVKKPKKVEAAGDEDVDIGDEMPMSEFP--PVEIEKDKDVAAGHASSSSGSSSSS 268
Query: 264 HSKSSTSS 271
S SS+SS
Sbjct: 269 SSGSSSSS 292
>TC8486 similar to GB|AAP68317.1|31711922|BT008878 At5g64430 {Arabidopsis
thaliana;}, partial (25%)
Length = 856
Score = 39.7 bits (91), Expect = 0.002
Identities = 36/140 (25%), Positives = 57/140 (40%), Gaps = 2/140 (1%)
Frame = +2
Query: 171 KLKIPKTRKRGATPTTSASQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASS 230
+ +IP TR+RG +TS ++ + + T PS + + + R SS
Sbjct: 56 RTQIPATRRRGRGRSTSRIRRHGRTSRVHTTTKPSSSAATAGR---------------SS 190
Query: 231 PLQDTTSIPVEMA--QAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMA 288
TTS P A ++ ++G SS++S SS + SSP S P R+
Sbjct: 191 HAPTTTSSPTSAATPRSSPSTGTPSSQRSSPSSPPSATLHRKSSPSSTSSPARISTP--- 361
Query: 289 VPRATWFSSKPPPIALSSSS 308
S P P +SS+
Sbjct: 362 --------SSPSPTTTTSST 397
>TC15534 weakly similar to GB|AAO63420.1|28950993|BT005356 At5g28040
{Arabidopsis thaliana;}, partial (8%)
Length = 937
Score = 39.3 bits (90), Expect = 0.002
Identities = 41/198 (20%), Positives = 78/198 (38%), Gaps = 11/198 (5%)
Frame = -3
Query: 134 IGQAEPIPRPTPQASPRVASATVEMEDDDDDDV-----ALADKLKIPKTRKRGATPTTSA 188
I A+ RP+P P AS++ E E++DD + A A++ ++ + + G++
Sbjct: 896 ITMAQKQKRPSPLEDPPTASSSSESEEEDDQPLSQVKHAAAEEEEL-SSEEEGSSEEEEE 720
Query: 189 SQKRAKGAGGSTVGNPSPAKSASQREEMQQGGDQGVDQQASSPLQDTTSIPVEMAQAKKT 248
+ A +T + P + + + Q + +P + S P + K
Sbjct: 719 DETTAPAPPAATTSHTKPPQPEPESDSATQSDSESDTDSDQAP---SASAPTPNPKVKPL 549
Query: 249 SGKRSSRKSGSSSSHHSKSSTSSSPLKASGPK------RVGMSKMAVPRATWFSSKPPPI 302
+ K + + +H K+ S +P K++ + G SK A +A S P
Sbjct: 548 ATKPMDQ----TQTHKPKAQPSPAPAKSAAKRAAESNANAGDSKRAKKKAV--DSAPAAA 387
Query: 303 ALSSSSSDLESSSSSLDS 320
A S + + S DS
Sbjct: 386 AGSDEEMEEDGKKSGNDS 333
>TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precursor (Atperox
P31) (ATP41) , partial (38%)
Length = 475
Score = 38.9 bits (89), Expect = 0.003
Identities = 29/95 (30%), Positives = 42/95 (43%)
Frame = +1
Query: 229 SSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMA 288
+SP T P + + RS+ S S+T+S+P A+ P
Sbjct: 142 NSPKSSATPSPTNKSPPPPPAPPRSA----------SSSTTASTPTAATPPSSSPQLPST 291
Query: 289 VPRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
P AT S+ P P S+SSS+L+ S+SL T S
Sbjct: 292 KPNATPTSTSPSPATPSTSSSELKPPSNSLAPTPS 396
>TC19039 similar to UP|Q8L7W5 (Q8L7W5) AT4g12110/F16J13_180, partial (61%)
Length = 763
Score = 38.5 bits (88), Expect = 0.003
Identities = 31/75 (41%), Positives = 38/75 (50%), Gaps = 2/75 (2%)
Frame = +2
Query: 248 TSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMS--KMAVPRATWFSSKPPPIALS 305
TS ++ S SSSS S S +SSS AS P + S + A P T S+ P A S
Sbjct: 254 TSSTATTFSSSSSSSPSSLSPSSSSRSNASPPSTITRSNPRFASPSVTCSSATRPSCACS 433
Query: 306 SSSSDLESSSSSLDS 320
SSSS +SS L S
Sbjct: 434 SSSSVPSNSSPILPS 478
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 38.1 bits (87), Expect = 0.004
Identities = 38/111 (34%), Positives = 50/111 (44%), Gaps = 15/111 (13%)
Frame = +1
Query: 228 ASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSS------PLKASGPKR 281
+SS ++S P + + +S SS S SSSS S SS+SSS P A GP
Sbjct: 295 SSSSSLSSSSTPSSSSSSPPSSSSSSSSSSSSSSSWSSPSSSSSSSPFSSGPAPAPGPPS 474
Query: 282 VGMSKMAV---------PRATWFSSKPPPIALSSSSSDLESSSSSLDSTES 323
++ + P + SS P P S SSS S SSSL S+ S
Sbjct: 475 GAFFELLLLFELLPYPAPSSDDKSSSPSPNGPSPSSS--SSPSSSLPSSPS 621
Score = 32.3 bits (72), Expect = 0.25
Identities = 36/118 (30%), Positives = 49/118 (41%), Gaps = 3/118 (2%)
Frame = +1
Query: 258 GSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSSSSSDLESSSSS 317
G S+S S SS+SSS +S + P ++ SS PP SSS SSSSS
Sbjct: 265 GCSASSSSSSSSSSS-----------LSSSSTPSSS--SSSPP------SSSSSSSSSSS 387
Query: 318 LDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECRDNLSNFP---PQDMQNRPSP 372
S+ S P + F+S P P ++ L +P D + PSP
Sbjct: 388 SSSSWSSPSSSSSSSPFSSGPAPAPGPPSGAFFELLLLFELLPYPAPSSDDKSSSPSP 561
>CB826981
Length = 591
Score = 37.4 bits (85), Expect = 0.008
Identities = 34/116 (29%), Positives = 43/116 (36%), Gaps = 19/116 (16%)
Frame = +3
Query: 341 WQHPGCEPYYTKECRDNLSNFPPQDMQNRPSPAREENIPHDEVQGGEDP----------- 389
WQ CE Y K+C +L N P +NR + EVQG + P
Sbjct: 96 WQGACCESYRRKKCLIDL-NLAPAVTENREQEGPSSQVVTQEVQGDQQPPVAQPPQDALP 272
Query: 390 --IIQPDQVI---GVMPPPKADVLASAHS---EPSPRQFQTSPQQPRLSPQRLQQD 437
+ QP +V V PP DV A E SPR F + R + R D
Sbjct: 273 PEVAQPPEVAQQPQVAQPPLIDVEAIDDDDVIESSPRAFAEAKNNSRRNRARAVVD 440
>TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial (7%)
Length = 841
Score = 36.6 bits (83), Expect = 0.013
Identities = 22/71 (30%), Positives = 31/71 (42%)
Frame = +1
Query: 366 MQNRPSPAREENIPHDEVQGGEDPIIQPDQVIGVMPPPKADVLASAHSEPSPRQFQTSPQ 425
+ N P P + + PH + Q P QP P P+ S+P P+ Q S
Sbjct: 13 LHNEPQPLQYQPQPHLQHQPQHHPHFQPH------PQPQLQ------SQPQPQHIQQSQP 156
Query: 426 QPRLSPQRLQQ 436
QP+ PQ +QQ
Sbjct: 157 QPQSQPQHIQQ 189
>TC16836 similar to GB|AAB68983.1|500656|YSCH9666 Yhr143wp {Saccharomyces
cerevisiae;} , partial (5%)
Length = 646
Score = 36.6 bits (83), Expect = 0.013
Identities = 31/84 (36%), Positives = 42/84 (49%), Gaps = 5/84 (5%)
Frame = +1
Query: 245 AKKTSGKRSSRKSGSSSSHHSK--SSTSSSPLKASGPKRVGMSKMAVP---RATWFSSKP 299
A SG+ SSR G+SSS + S SSS +G R G + +P R+T + P
Sbjct: 169 ASTVSGRASSRSPGTSSSRSCRWCRSVSSS*WTFTGSTRRGQAATVIPALRRSTSVTRNP 348
Query: 300 PPIALSSSSSDLESSSSSLDSTES 323
A ++ SS L SSS+ ST S
Sbjct: 349 S*RASATRSSSLPRSSSTGFSTPS 420
>TC8946 homologue to GB|AAM66972.1|21617922|AY088650 pyridoxine
biosynthesis protein-like {Arabidopsis thaliana;} ,
partial (45%)
Length = 503
Score = 36.2 bits (82), Expect = 0.017
Identities = 30/116 (25%), Positives = 46/116 (38%), Gaps = 3/116 (2%)
Frame = +2
Query: 226 QQASSPLQDTTSIPVEMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMS 285
QQ L+ + S + + S+ + S + S TSS+P K + PK+
Sbjct: 74 QQQWKDLESSLSTATAPQSPRPRNPPSPSKSASLRCSAAASSWTSSTPNKPASPKK---- 241
Query: 286 KMAVPRATWFSSKPPPIALSSSSS---DLESSSSSLDSTESDPVWDKLTFYFNSKP 338
P +W SS PP + ++S SSS + P W K +SKP
Sbjct: 242 --PAPAPSWPSSVSPPTSAPKAASLA*ATLSSSRKSNKPSPSPSWPKPASAISSKP 403
>TC17998 homologue to UP|Q8UKS4 (Q8UKS4) IS21 family transposase, partial
(14%)
Length = 564
Score = 35.8 bits (81), Expect = 0.022
Identities = 18/29 (62%), Positives = 22/29 (75%)
Frame = -3
Query: 622 RGSEDLKIRGSEDQKIRGSEDLKIRGSED 650
RGSE+L RGSE+ RGSE+L RGSE+
Sbjct: 193 RGSENLSPRGSENLSPRGSENLSPRGSEN 107
Score = 32.7 bits (73), Expect = 0.19
Identities = 17/30 (56%), Positives = 21/30 (69%)
Frame = -3
Query: 614 RSSEDQKIRGSEDLKIRGSEDQKIRGSEDL 643
R SE+ RGSE+L RGSE+ RGSE+L
Sbjct: 193 RGSENLSPRGSENLSPRGSENLSPRGSENL 104
Score = 29.6 bits (65), Expect = 1.6
Identities = 18/39 (46%), Positives = 23/39 (58%), Gaps = 10/39 (25%)
Frame = -3
Query: 622 RGSEDLKIRGSEDQKI----------RGSEDLKIRGSED 650
RGSE+L RGSE+ + RGSE+L RGSE+
Sbjct: 349 RGSENLSPRGSENLSLLVADRL*LESRGSENLSPRGSEN 233
>TC19223 similar to UP|Q940C3 (Q940C3) AT3g27530/MMJ24_7, partial (13%)
Length = 525
Score = 34.7 bits (78), Expect = 0.050
Identities = 24/74 (32%), Positives = 35/74 (46%)
Frame = +2
Query: 247 KTSGKRSSRKSGSSSSHHSKSSTSSSPLKASGPKRVGMSKMAVPRATWFSSKPPPIALSS 306
KT K S SSS+ + S++S +P + S +A+ +TW S P + S
Sbjct: 233 KTEIKVHHSSSSSSSTSSAASTSSPTPSSSWSTSSPSSSNLALSLSTWLGSGPRHTSRSF 412
Query: 307 SSSDLESSSSSLDS 320
SS L +SSL S
Sbjct: 413 SSPSLSF*ASSLAS 454
>TC15341 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (33%)
Length = 1002
Score = 32.7 bits (73), Expect = 0.19
Identities = 49/221 (22%), Positives = 82/221 (36%), Gaps = 9/221 (4%)
Frame = +1
Query: 241 EMAQAKKTSGKRSSRKSGSSSSHHSKSSTSSSP-----LKASGPKRVGMSKMAVPRATWF 295
E+A+ +K K + SSSS HS+S+ SP KA ++A+
Sbjct: 406 ELAETQKELAKLQLAQKESSSSSHSQSNEDKSPSTTDSKKADNASDASNQQLALALPNQI 585
Query: 296 SSKPPPIALSSSSSDLESSSSSLDSTESDPVWDKLTFYFNSKPIAWQHPGCEPYYTKECR 355
+ +P P A + ++ + ++ P +Y S P+ P T+
Sbjct: 586 APQPQPAA-----AQPQAPAPNVTQAPQQP-----AYYIPSTPL--------PGNTQTVA 711
Query: 356 DNLSN-FPPQDMQNRPSPAREENIPHDEVQGGEDPIIQP-DQVIGVMPPPKADVLASAHS 413
N + P D Q R ++Q QP G PPP +
Sbjct: 712 QLPQNQYLPPDQQYRT----------PQMQDMSRVAPQPCTNXSGXSPPPPVQQYPQ-YQ 858
Query: 414 EPSPRQFQTSPQQ--PRLSPQRLQQDARAALLSPHAKGGSA 452
+P +Q Q QQ P+ PQ++Q ++ SP + S+
Sbjct: 859 QPQQQQQQQQQQQQWPQQLPQQVQPTQPPSMQSPQIRPPSS 981
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,938,377
Number of Sequences: 28460
Number of extensions: 191541
Number of successful extensions: 2338
Number of sequences better than 10.0: 182
Number of HSP's better than 10.0 without gapping: 1592
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1975
length of query: 653
length of database: 4,897,600
effective HSP length: 96
effective length of query: 557
effective length of database: 2,165,440
effective search space: 1206150080
effective search space used: 1206150080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0545.2


