

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
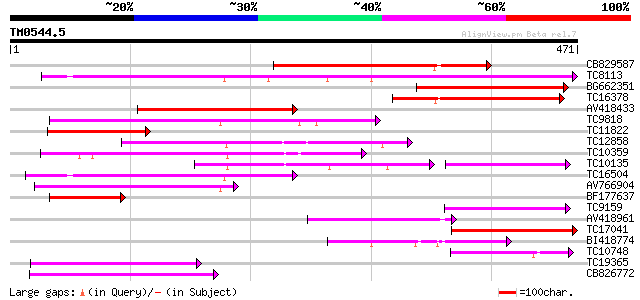
Query= TM0544.5
(471 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB829587 194 2e-50
TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter li... 167 4e-42
BG662351 150 6e-37
TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22... 145 1e-35
AV418433 142 9e-35
TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, ... 120 6e-28
TC11822 similar to UP|P93051 (P93051) ORFc protein (Fragment), p... 116 9e-27
TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter,... 111 2e-25
TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partia... 105 1e-23
TC10135 95 3e-20
TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporte... 87 4e-18
AV766904 84 6e-17
BF177637 78 4e-15
TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose tr... 77 8e-15
AV418961 75 3e-14
TC17041 homologue to GB|AAL25568.1|16648753|AY058152 AT5g16150/T... 72 2e-13
BI418774 72 3e-13
TC10748 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter,... 68 4e-12
TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter l... 67 6e-12
CB826772 66 1e-11
>CB829587
Length = 548
Score = 194 bits (494), Expect = 2e-50
Identities = 98/185 (52%), Positives = 133/185 (70%), Gaps = 4/185 (2%)
Frame = +2
Query: 220 ALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGG 279
AL+RLRGK+ D+ EA EI D ETLQ ++ ++ LF +Y+ S+ IGVGLM+ QQ G
Sbjct: 2 ALRRLRGKDVDISHEATEILDSIETLQSLPKSKLLDLFHTRYVRSVVIGVGLMVCQQSVG 181
Query: 280 INGIVFYANSIFVSAGFSE-SIGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCL 338
INGI FY FV+AG S GTIA +++P T+LG +LMDKSGRRPL+ +S+ GT L
Sbjct: 182 INGIGFYTAETFVAAGVSSGKAGTIAYACIQVPFTILGAILMDKSGRRPLITVSSGGTFL 361
Query: 339 GCFLAALAFLLQG---LHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVK 395
GCF+ +AF L+ L EW P LA+ GVL+Y+ +YS+G+G++PWVIMSE+FPI++K
Sbjct: 362 GCFITGIAFFLKDQGLLLEW---VPTLAIGGVLIYIAAYSIGLGSVPWVIMSEVFPIHIK 532
Query: 396 GSAGS 400
G+AGS
Sbjct: 533 GTAGS 547
>TC8113 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (93%)
Length = 1865
Score = 167 bits (422), Expect = 4e-42
Identities = 119/474 (25%), Positives = 212/474 (44%), Gaps = 29/474 (6%)
Frame = +2
Query: 27 ATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVS 86
A + ++++V+++ Y G G I D+ ++ Q + IL I A++G + +
Sbjct: 122 ACVIVASMVSIISGYDTGVMSG----ALLFIKEDIGISDTQQEVLAGILNICALVGCLAA 289
Query: 87 GKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAE 146
GK +DY GRR + + + ++G + + + L GR + G G+G PVY AE
Sbjct: 290 GKTSDYIGRRYTIFLASILFLVGAVFMGYGPNFAILMFGRCVCGLGVGFALTTAPVYSAE 469
Query: 147 ITPKNLRGGFTTMHQLMICCGMSLTYLIGAF-------VNWRILALIGAIPCFAQLLSLP 199
++ + RG T++ ++ I G+ + Y+ F + WR++ + AIP L +
Sbjct: 470 LSSASTRGFLTSLPEVCIGLGIFIGYISNYFLGKLALTLGWRLMLGLAAIPSLGLALGIL 649
Query: 200 FIPDSPRWLAKVGH-------LKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEAS 252
+P+SPRWL G L E + + + D+ A + +Q + S
Sbjct: 650 TMPESPRWLVMQGRLGCAKKVLLEVSNTTEEAELRFRDIIVSAGFDEKCNDEFVKQPQKS 829
Query: 253 IIGLFQWKYM---------TSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES---- 299
G WK + L VG+ + GI ++ Y IF AG +
Sbjct: 830 HHGEGVWKELFLRPTPPVRRMLIAAVGIHFFEHATGIEAVMLYGPRIFKKAGVTTKDRLL 1009
Query: 300 IGTIAMVAVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVS 359
+ TI KI + L+D+ GRR LL +S AG G + + + K V
Sbjct: 1010LATIGTGLTKITFLTISTFLLDRVGRRRLLQISVAGMIFGLTILGFSLTMVEYSSEKLVW 1189
Query: 360 PI-LALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAF- 417
+ L++V YV +++G+ + WV SEIFP+ ++ S+ N + +S +F
Sbjct: 1190ALSLSIVATYTYVAFFNVGLAPVTWVYSSEIFPLRLRAQGNSIGVAVNRGMNAAISMSFI 1369
Query: 418 NFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHSTKRLI 471
+ + + G FF+F+ + +F +PETKG+ LEE++ S S + +
Sbjct: 1370SIYKAITIGGAFFLFAGMSVVAWVFFYFCLPETKGKALEEMEMVFSKKSDDKTV 1531
>BG662351
Length = 407
Score = 150 bits (378), Expect = 6e-37
Identities = 66/126 (52%), Positives = 95/126 (75%)
Frame = +1
Query: 339 GCFLAALAFLLQGLHEWKGVSPILALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSA 398
G L A+AF L+ G P LA+ G+LVYVGS+++GMGA+PWV+MSEIFP+N+KG A
Sbjct: 16 GSILTAVAFYLKAQEISVGAVPGLAVTGILVYVGSFAIGMGAVPWVVMSEIFPVNIKGQA 195
Query: 399 GSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFSSICGFTVLFVAKLVPETKGRTLEEI 458
GSL T NW +W+ SY FNFLMSWS+ GTF ++++I +L + +VPETKG++LE++
Sbjct: 196 GSLATLVNWFGAWLCSYTFNFLMSWSTYGTFILYAAINALGILSIVVVVPETKGKSLEQL 375
Query: 459 QASLSS 464
QA++++
Sbjct: 376 QAAINA 393
>TC16378 similar to GB|AAM70554.1|21700861|AY124845 At1g75220/F22H5_6
{Arabidopsis thaliana;}, partial (31%)
Length = 738
Score = 145 bits (366), Expect = 1e-35
Identities = 75/148 (50%), Positives = 105/148 (70%), Gaps = 5/148 (3%)
Frame = +2
Query: 319 LMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGL-----HEWKGVSPILALVGVLVYVGS 373
L+DKSGRR LLLLS++ + + ++AF L+G H +K + I+++VG++V V
Sbjct: 11 LVDKSGRRLLLLLSSSIMTVSLVVVSIAFYLEGTVSEDSHLYK-ILGIVSVVGLVVMVIG 187
Query: 374 YSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSSTGTFFIFS 433
+SLG+G IPWVIMSEI P+++KG AGS T NWL SW ++ N L++WSS GTF +++
Sbjct: 188 FSLGLGPIPWVIMSEILPVSIKGLAGSTATMANWLTSWAITMTANLLLTWSSGGTFTMYT 367
Query: 434 SICGFTVLFVAKLVPETKGRTLEEIQAS 461
+ FTV+F A VPETKGRTLEEIQ+S
Sbjct: 368 VVAAFTVVFTALWVPETKGRTLEEIQSS 451
>AV418433
Length = 401
Score = 142 bits (359), Expect = 9e-35
Identities = 71/133 (53%), Positives = 96/133 (71%)
Frame = +3
Query: 107 ILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICC 166
+ GWLVI FS+ L +GR+ G G+G+ SYVVPV++AEI PK LRG TT++Q M+
Sbjct: 3 VAGWLVIYFSQGPVLLDIGRVATGYGMGVFSYVVPVFIAEIAPKELRGALTTLNQFMLVS 182
Query: 167 GMSLTYLIGAFVNWRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRG 226
MS++++IG ++WR LAL G IP LL L FIP+SPRWLAK G ++ ++ALQ LRG
Sbjct: 183 AMSVSFVIGNVLSWRALALTGLIPTAVLLLGLFFIPESPRWLAKRGCREDFEAALQILRG 362
Query: 227 KNADVHQEAAEIR 239
K+AD+ QEA EI+
Sbjct: 363 KDADISQEAEEIQ 401
>TC9818 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(62%)
Length = 1162
Score = 120 bits (300), Expect = 6e-28
Identities = 83/298 (27%), Positives = 145/298 (47%), Gaps = 23/298 (7%)
Frame = +2
Query: 34 LVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVADYA 93
++A + S G IG S I DL ++ + I I+ + ++IG+ ++G+ +D+
Sbjct: 251 MLASMTSILLGYDIGVMSGAAIYIKRDLKVSDVKIEILLGIINLYSLIGSGLAGRTSDWI 430
Query: 94 GRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLR 153
GRR + F+ +G L++ FS W+L GR + G GIG + PVY AE++P + R
Sbjct: 431 GRRYTIVFAGAIFFVGALLMGFSPNYWFLMFGRFIAGIGIGYALMIAPVYTAEVSPASSR 610
Query: 154 GGFTTMHQLMICCGMSLTYL-------IGAFVNWRILALIGAIPCFAQLLSLPFIPDSPR 206
G T+ ++ I G+ L Y+ + V WR++ +GA+P + + +P+SPR
Sbjct: 611 GFLTSFPEVFINGGILLGYISNFAFSKLSLKVGWRMMLGVGALPSVILGVGVLAMPESPR 790
Query: 207 WLAKVGHLKESDSALQRLRGKNADVHQEAAEIR-----------DYTETLQRQTEASI-- 253
WL G L ++ L + + A+I+ D E +R T +
Sbjct: 791 WLVMRGRLGDAIKVLNKTSDSPEEAQLRLADIKRAAGIPESCTDDVVEVSKRSTGEGVWK 970
Query: 254 -IGLFQWKYMTSLTI-GVGLMILQQFGGINGIVFYANSIFVSAGF-SESIGTIAMVAV 308
+ L+ + + I +G+ QQ GI+ +V Y+ +IF AG S++ +A VAV
Sbjct: 971 ELFLYPTPAIRHIVIAALGIHFFQQASGIDAVVLYSPTIFEKAGIKSDTDKLLATVAV 1144
>TC11822 similar to UP|P93051 (P93051) ORFc protein (Fragment), partial
(74%)
Length = 419
Score = 116 bits (290), Expect = 9e-27
Identities = 56/86 (65%), Positives = 68/86 (78%)
Frame = +3
Query: 32 STLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVAD 91
+T VAV GSY FGA IGYSSPTQ AI DLNL++A++S+FGSI+T GAMIGAI SG +AD
Sbjct: 156 TTFVAVCGSYEFGAGIGYSSPTQDAIRKDLNLSLAEYSLFGSIVTFGAMIGAITSGLIAD 335
Query: 92 YAGRRVAMGFSQLFCILGWLVITFSK 117
+ GR+ AM S FC+ GWLVI FS+
Sbjct: 336 FIGRKGAMRVSSAFCVAGWLVIYFSQ 413
>TC12858 weakly similar to UP|Q9ZS76 (Q9ZS76) Hexose transporter, partial
(51%)
Length = 882
Score = 111 bits (278), Expect = 2e-25
Identities = 78/251 (31%), Positives = 118/251 (46%), Gaps = 10/251 (3%)
Frame = +3
Query: 94 GRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYVVPVYVAEITPKNLR 153
G R M LF + G L + W L VGRLL+G GIG + VP+Y++E+ P R
Sbjct: 36 GMRATMIVGGLFFVSGALFNGLADGIWMLIVGRLLLGFGIGCANQSVPIYLSEMAPYKYR 215
Query: 154 GGFTTMHQLMICCGMSLTYLIGAFV-------NWRILALIGAIPCFAQLLSLPFIPDSPR 206
GG QL I G+ + L + WR+ +GA+P ++ +PDSP
Sbjct: 216 GGLNMCFQLSITIGIFVANLFNYYFAKILNGQGWRLSLGLGAVPAVIFVVGSLCLPDSPS 395
Query: 207 WLAKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTSLT 266
L G + + L ++RG D+ E +I +E L+ + L + KY L
Sbjct: 396 SLVARGRHEAARQELVKIRG-TTDIEAELKDIITASEALE-NVKHPWKTLLERKYRPQLV 569
Query: 267 IGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAV---KIPVTVLGVLLMDKS 323
V + QQF G+N I FYA +F + GF + ++ V + K T++ + ++DK
Sbjct: 570 FAVCIPFFQQFTGLNVITFYAPILFRTIGFGPTASLMSAVIIGSFKPVSTLISIFVVDKF 749
Query: 324 GRRPLLLLSAA 334
GRR L L A
Sbjct: 750 GRRTLFLEGGA 782
>TC10359 similar to UP|Q94AZ2 (Q94AZ2) AT5g26340/F9D12_17, partial (90%)
Length = 1099
Score = 105 bits (263), Expect = 1e-23
Identities = 78/299 (26%), Positives = 133/299 (44%), Gaps = 28/299 (9%)
Frame = +2
Query: 26 TATLTLSTLVAVLGSYAFGAAIGYSSPTQSA------IMPDL-NLTVAQ----------- 67
T + +S ++A G FG +G S S PD+ TV +
Sbjct: 200 TPIVIISCILAATGGLMFGYDVGVSGGVTSMPHFLHKFFPDVYKKTVLEKELDSNYCKYD 379
Query: 68 ---FSIFGSILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYV 124
+F S L + ++ + GRR+ M + +F I G + ++ L V
Sbjct: 380 NQGLQLFTSSLYLAGLVSTFFASYTTRTLGRRMTMLIAGVFFIAGVVFNVTAQNLAMLIV 559
Query: 125 GRLLVGCGIGLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAFVN----- 179
GR+L+GCG+G + VP++++EI P +RG + QL I G+ LI N
Sbjct: 560 GRILLGCGVGFANQAVPLFLSEIAPSRIRGALNILFQLNITIGILFANLINYGTNKIKGG 739
Query: 180 --WRILALIGAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAE 237
WR+ + IP + + D+P L + G +E + L+++RG + +V E E
Sbjct: 740 WGWRLSLGLAGIPAVLLTVGALLVVDTPNSLIERGRTEEGKAVLKKIRGTD-NVEPEFLE 916
Query: 238 IRDYTETLQRQTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGF 296
+ + + ++ + L + + + I + L I QQF GIN I+FYA +F + GF
Sbjct: 917 LVE-ASRVAKEVKDPFRNLLKRRNRPQIIISIALQIFQQFTGINAIMFYAPVLFNTLGF 1090
>TC10135
Length = 1501
Score = 94.7 bits (234), Expect = 3e-20
Identities = 63/213 (29%), Positives = 109/213 (50%), Gaps = 13/213 (6%)
Frame = +2
Query: 154 GGFTTMHQLMICCGMSLTYLIG-AFVN----WRILALIGAIPCFAQLLSLPFIPDSPRWL 208
G +++ +I G L+YLI AF N WR + + A P Q++ + +P+SPRWL
Sbjct: 2 GALVSLNSFLITGGQFLSYLINLAFTNAPGTWRWMLGVAAAPAIIQIVLMLSLPESPRWL 181
Query: 209 AKVGHLKESDSALQRLRGKNADVHQEAAEIRDYTETLQRQTEASIIGLFQWKYMTS---- 264
+ G +ES S L+++ DV E +++ E+ +++ S I + T+
Sbjct: 182 YRKGREEESKSILKKIYAPE-DVDAEIEALKESVESEIEESKTSKISMMTLLKTTTVRRG 358
Query: 265 LTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMVAVKIPV----TVLGVLLM 320
L G+GL QQF GIN +++Y+ +I AGF+ + + + + + ++L + +
Sbjct: 359 LYAGMGLQFFQQFVGINTVMYYSPTIVQLAGFASNRTALLLSLITSGLNAFGSILSIYFI 538
Query: 321 DKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLH 353
DK+GR+ L L+S G L L +AF LH
Sbjct: 539 DKTGRKKLALISLCGVVLSLALLTVAFRESELH 637
Score = 79.7 bits (195), Expect = 9e-16
Identities = 41/105 (39%), Positives = 60/105 (57%), Gaps = 1/105 (0%)
Frame = +2
Query: 363 ALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMS 422
ALVG+ +Y+ ++S GMG +PWVI SEI+P +G G + + W+ + IVS +F L
Sbjct: 893 ALVGLALYIITFSPGMGTVPWVINSEIYPFRYRGVCGGIASTTVWISNLIVSQSFLSLTQ 1072
Query: 423 WSSTG-TFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHS 466
T TF +F + + FV VPETKG +EE++ L S
Sbjct: 1073TIGTAWTFMMFGILAVVAIFFVIVFVPETKGVPMEEVEKMLEQRS 1207
>TC16504 weakly similar to UP|Q84KI7 (Q84KI7) Sorbitol transporter, partial
(37%)
Length = 903
Score = 87.4 bits (215), Expect = 4e-18
Identities = 57/233 (24%), Positives = 107/233 (45%), Gaps = 7/233 (3%)
Frame = +2
Query: 14 FNDEEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGS 73
F D ++ A++ ++++ L Y G I DL L+ Q
Sbjct: 53 FGDSHKRLNMYACASVVAASVIFALYGYVMAVMTG----VLLFIKEDLQLSDLQVQFSVG 220
Query: 74 ILTIGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGI 133
+L + A+ + +G++ADY GRR + + + ++G +++ + L +G + G G+
Sbjct: 221 LLHMSALPACMTAGRIADYIGRRYTIILTAIIFLVGSVLMGYGPSYPILMIGLSISGIGL 400
Query: 134 GLLSYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYLIGAF-------VNWRILALI 186
G V PVY AEI+P + RG T+ ++ G++L Y+ F + WR++ ++
Sbjct: 401 GFSLIVAPVYCAEISPPSHRGFLTSFLEVSTNAGIALGYVSNYFFGKLSIRLGWRLMLVV 580
Query: 187 GAIPCFAQLLSLPFIPDSPRWLAKVGHLKESDSALQRLRGKNADVHQEAAEIR 239
A+P A + + +SPRWL G + E+ L + + Q EI+
Sbjct: 581 HAVPSLALVALMLNWVESPRWLVMQGRVGEARKVLLLVSNTKEEAEQRLKEIK 739
>AV766904
Length = 611
Score = 83.6 bits (205), Expect = 6e-17
Identities = 52/178 (29%), Positives = 91/178 (50%), Gaps = 8/178 (4%)
Frame = +3
Query: 21 QQRPFTATLTLS-TLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGA 79
Q++P L+ ++A + S G IG S I DL ++ Q I I+ I +
Sbjct: 63 QKKPKRNKFALACAILASMTSILLGYDIGVMSGAVIYIKRDLKISDVQIEILSGIMNIYS 242
Query: 80 MIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLSYV 139
IG+ ++G+ +D+ GRR + + L +G +++ FS +L GR + G GIG +
Sbjct: 243 PIGSYIAGRTSDWIGRRYTIVLAGLIFFVGAILMGFSPNYAFLMFGRFVAGVGIGFAFLI 422
Query: 140 VPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYL-------IGAFVNWRILALIGAIP 190
PVY +EI+P RG T++ ++ + G+ + Y+ + + WR++ IGAIP
Sbjct: 423 APVYTSEISPTLSRGFLTSLPEVFLNAGILIGYISNYTFSKLPLRLGWRLMLGIGAIP 596
>BF177637
Length = 480
Score = 77.8 bits (190), Expect = 4e-15
Identities = 34/63 (53%), Positives = 49/63 (76%)
Frame = +2
Query: 34 LVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTIGAMIGAIVSGKVADYA 93
L+ LG FG GY+SPTQ+AI+ DL L+V++FS+FGS+ +GAM+GAI SG++A+Y
Sbjct: 272 LIVALGPIQFGFTAGYTSPTQAAIISDLGLSVSEFSLFGSLSNVGAMLGAIASGQIAEYI 451
Query: 94 GRR 96
GR+
Sbjct: 452 GRK 460
>TC9159 similar to UP|HXT4_YEAST (P32467) Low-affinity glucose transporter
HXT4 (Low-affinity glucose transporter LGT1), partial
(5%)
Length = 728
Score = 76.6 bits (187), Expect = 8e-15
Identities = 39/106 (36%), Positives = 62/106 (57%), Gaps = 1/106 (0%)
Frame = +3
Query: 362 LALVGVLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLM 421
LAL G+ +Y+ +S GMG +PWV+ SEI+P+ +G G + + W+ + +VS +F L
Sbjct: 117 LALGGLALYIIFFSPGMGTVPWVVNSEIYPLRYRGICGGIASTTVWVSNLVVSQSFLSLT 296
Query: 422 SWSSTG-TFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHS 466
T TF +F I + FV VPETKG +EE+++ L+ +
Sbjct: 297 QTIGTAWTFMMFGIIAIIGIFFVLIFVPETKGVPMEEVESMLNKRA 434
>AV418961
Length = 394
Score = 74.7 bits (182), Expect = 3e-14
Identities = 42/125 (33%), Positives = 69/125 (54%), Gaps = 1/125 (0%)
Frame = +3
Query: 248 QTEASIIGLFQWKYMTSLTIGVGLMILQQFGGINGIVFYANSIFVSAGFSESIGTIAMV- 306
+ EA LF +Y +++G L + QQ GIN +V+Y+ S+F SAG + + A+V
Sbjct: 21 EPEAGWFDLFSTRYWKVVSVGAALFLFQQLAGINAVVYYSTSVFRSAGIASDVAASALVG 200
Query: 307 AVKIPVTVLGVLLMDKSGRRPLLLLSAAGTCLGCFLAALAFLLQGLHEWKGVSPILALVG 366
A + T + LMD+ GR+ LL+ S +G L +L+F + L + G LA++G
Sbjct: 201 ASNVFGTAVASSLMDRQGRKSLLITSFSGMAASMLLLSLSFTWKVLTPYSGT---LAVLG 371
Query: 367 VLVYV 371
++YV
Sbjct: 372 TVLYV 386
>TC17041 homologue to GB|AAL25568.1|16648753|AY058152 AT5g16150/T21H19_70
{Arabidopsis thaliana;} , partial (18%)
Length = 632
Score = 72.0 bits (175), Expect = 2e-13
Identities = 38/106 (35%), Positives = 67/106 (62%), Gaps = 2/106 (1%)
Frame = +2
Query: 368 LVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVS-YAFNFLMSWSST 426
++YV S+SLG G +P +++ EIF ++ A SL +W+ ++++ Y + + + +
Sbjct: 5 VLYVLSFSLGAGPVPALLLPEIFASRIRAKAVSLSLGMHWISNFVIGLYFLSVVNKFGIS 184
Query: 427 GTFFIFSSICGFTVLFVAKLVPETKGRTLEEIQASLSSHST-KRLI 471
+ FS++C VL++A V ETKGR+LEEI+ +LSS S K+L+
Sbjct: 185 SVYLGFSAVCVLAVLYIAGNVVETKGRSLEEIEHALSSSSA*KKLV 322
>BI418774
Length = 554
Score = 71.6 bits (174), Expect = 3e-13
Identities = 48/163 (29%), Positives = 82/163 (49%), Gaps = 10/163 (6%)
Frame = +3
Query: 265 LTIGVGLMILQQFGGINGIVFYANSIFVSAGFSES----IGTIAMVAVKIPVTVLGVLLM 320
L +G+ + QQ GI GI Y+ +F G + + T+ + + T + L+
Sbjct: 72 LIAAIGVHVFQQICGIEGICIYSPRVFEKTGITSKSNLLLATVGLGISQAVFTFISAFLL 251
Query: 321 DKSGRRPLLLLSAAG---TCLGCFLAALAFLLQGLHE---WKGVSPILALVGVLVYVGSY 374
D+ GRR LLL+S+ G T LG L+ +F++Q E W G+S +V + ++V
Sbjct: 252 DRVGRRILLLISSGGVVVTLLG--LSFCSFIMQQSKEEPLW-GIS--FTIVAIYIFVAFV 416
Query: 375 SLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAF 417
++G+G + WV SEIFP+ ++ + N L + V +F
Sbjct: 417 AIGIGPVTWVYSSEIFPLRLRAQGLGVCVLVNRLANVAVLTSF 545
>TC10748 similar to UP|Q7XA50 (Q7XA50) Sorbitol-like transporter, partial
(21%)
Length = 550
Score = 67.8 bits (164), Expect = 4e-12
Identities = 37/105 (35%), Positives = 61/105 (57%), Gaps = 3/105 (2%)
Frame = +2
Query: 367 VLVYVGSYSLGMGAIPWVIMSEIFPINVKGSAGSLVTFFNWLCSWIVSYAFNFLMSWSST 426
VL YV ++S+G G I WV SEIFP+ ++ ++ N + S ++S F L +
Sbjct: 5 VLSYVATFSIGAGPITWVYSSEIFPLRLRAQGCAMGVVVNRVTSGVISMTFLSLSKGITI 184
Query: 427 -GTFFIFS--SICGFTVLFVAKLVPETKGRTLEEIQASLSSHSTK 468
G FF+F +ICG+ +F ++PET+G+TLE+++ S +K
Sbjct: 185 GGAFFLFGGIAICGW--IFFYTMLPETRGKTLEDMEGSFGQSKSK 313
>TC19365 weakly similar to UP|O23213 (O23213) Sugar transporter like
protein, partial (24%)
Length = 441
Score = 67.0 bits (162), Expect = 6e-12
Identities = 40/142 (28%), Positives = 71/142 (49%)
Frame = +2
Query: 18 EQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILTI 77
E+ +++ T L L +++L FG G + I +L + Q + G IL +
Sbjct: 11 EKSRRKLNTVCLCLRDGLSLLYLAIFGYVTGVMAGALLFIKEELGINDMQVQLLGGILNV 190
Query: 78 GAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLLS 137
A+ +V+G+ +DY GRR + S + +LG +++ + L +GR G G+GL
Sbjct: 191 CALPACMVAGRTSDYVGRRYTIILSAVILLLGSILMGYGPSFPILMIGRCTAGLGVGLAL 370
Query: 138 YVVPVYVAEITPKNLRGGFTTM 159
+ PVY EI+ + RG T++
Sbjct: 371 IIAPVYSTEISLPSNRGFLTSL 436
>CB826772
Length = 490
Score = 66.2 bits (160), Expect = 1e-11
Identities = 39/157 (24%), Positives = 73/157 (45%)
Frame = +1
Query: 17 EEQQQQRPFTATLTLSTLVAVLGSYAFGAAIGYSSPTQSAIMPDLNLTVAQFSIFGSILT 76
E + R +S + A + S +G +G + I DL ++ Q I
Sbjct: 19 ENGETNRRLNLYACVSVMAASILSAIYGYEVGVMTGALLFIKEDLQISDLQLQFLVGIFH 198
Query: 77 IGAMIGAIVSGKVADYAGRRVAMGFSQLFCILGWLVITFSKVAWWLYVGRLLVGCGIGLL 136
+ + ++ +G+++DY GRR + + + + G ++ +S L +G + G G G
Sbjct: 199 MCGLPASMAAGRISDYIGRRYTIILASIAFLSGSILTGYSPSYLILMIGNFISGIGAGFT 378
Query: 137 SYVVPVYVAEITPKNLRGGFTTMHQLMICCGMSLTYL 173
V P+Y AEI+P + RG T++ + I G+ L Y+
Sbjct: 379 LIVAPLYCAEISPPSHRGSLTSLTEFSINIGILLGYM 489
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,456,010
Number of Sequences: 28460
Number of extensions: 124556
Number of successful extensions: 1061
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 1019
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1035
length of query: 471
length of database: 4,897,600
effective HSP length: 94
effective length of query: 377
effective length of database: 2,222,360
effective search space: 837829720
effective search space used: 837829720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0544.5


