

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0541.9
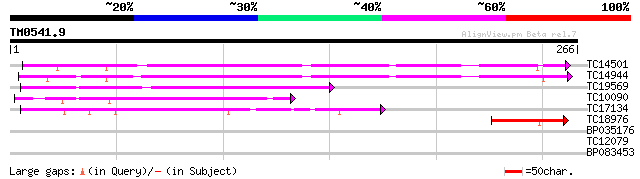
(266 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14501 similar to UP|LEC_LOTTE (P19664) Anti-H(O) lectin (LTA),... 160 2e-40
TC14944 similar to UP|LEC_LOTTE (P19664) Anti-H(O) lectin (LTA),... 140 2e-34
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 100 3e-22
TC10090 weakly similar to UP|Q9ZWP6 (Q9ZWP6) Lectin, partial (43%) 92 1e-19
TC17134 weakly similar to UP|LEC_BOWMI (P42088) Lectin (Agglutin... 57 2e-09
TC18976 similar to UP|LCB2_ROBPS (Q42372) Bark agglutinin I, pol... 40 5e-04
BP035176 39 0.001
TC12079 27 3.6
BP083453 26 8.0
>TC14501 similar to UP|LEC_LOTTE (P19664) Anti-H(O) lectin (LTA), partial
(86%)
Length = 1066
Score = 160 bits (406), Expect = 2e-40
Identities = 102/263 (38%), Positives = 149/263 (55%), Gaps = 6/263 (2%)
Frame = +1
Query: 7 FVILISLFMVAHNVN-SETFTFPGFQLPNTKDITFEGDA--FASNGVLELTKTAYGVPLP 63
F++ F++A NVN + +F + + + +GDA +A + T + G
Sbjct: 79 FLLACLFFLLAINVNLAAAISFNFTEFTDDGSLIIQGDAKIWADGSLALPTDPSVGF--- 249
Query: 64 SSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNS 123
++ RA +A PV +WD TG +A+F TSF+ ++ DG+ FF+APF ++IPK S
Sbjct: 250 -TTSRALYATPVPIWDSTTGNVASFVTSFSFIIKDFEDYNPADGLVFFLAPFGTEIPKES 426
Query: 124 TGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTWDPFTSHIGIDINSIISSKTVPWRTGNS 183
TGG G+ + AF NQIVAVEFDT+ + WD HIGID+NS+IS KTVPW N
Sbjct: 427 TGGRFGIINGKDAF----NQIVAVEFDTFINPWDSSPRHIGIDVNSLISLKTVPW---NK 585
Query: 184 LSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVRVGFS 243
++ S T+ Y+ TKTLSVLV + Q +++S +DL+ +LPE V VGFS
Sbjct: 586 VAGSLEKVTIIYDSQTKTLSVLVIHENGQ--------ISTISQEIDLKVVLPEEVSVGFS 741
Query: 244 GAT---GQLVEQHRIHVWDFKSS 263
T G+ E+H I+ W F S+
Sbjct: 742 ATTTSGGR--ERHDIYSWSFTST 804
>TC14944 similar to UP|LEC_LOTTE (P19664) Anti-H(O) lectin (LTA), partial
(85%)
Length = 1155
Score = 140 bits (354), Expect = 2e-34
Identities = 94/266 (35%), Positives = 144/266 (53%), Gaps = 6/266 (2%)
Frame = +2
Query: 5 SLFVILISLFMV----AHNVNSETFTFPGFQLPNTKDITFEGDA-FASNGVLELTKTAYG 59
+L V+L SL ++ ++V +F F F + + +GDA ++G L L
Sbjct: 62 TLKVVLASLLLLLTIKVNSVEGVSFNFTKFT--DDGSLILQGDAKIWTDGRLALPSDPL- 232
Query: 60 VPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKI 119
+ ++ R +A PV +WD TG++A+F +SF+ V DG+ FF+AP+ + I
Sbjct: 233 --IGKTTSRVLYATPVPIWDSTTGSVASFVSSFSFVVSDVPGYKISDGLVFFLAPWGTTI 406
Query: 120 PKNSTGGFLGLFSKDSAFDSYQNQIVAVEFDTYADTWDPFTSHIGIDINSIISSKTVPWR 179
P NS G LG+ + + + NQ VAVEFD+Y +T DP HIGID+NS++S V W
Sbjct: 407 PPNSEGKNLGILDEKNGY----NQFVAVEFDSYNNTRDPTFRHIGIDVNSVMSMNLVKW- 571
Query: 180 TGNSLSTSAAFATVSYEPVTKTLSVLVKQIDQQKGLKFSTITTSLSFVVDLRTILPEWVR 239
N +S + A + Y+ TKTLSV+V + Q T+++ +DL+ +LP+ V
Sbjct: 572 --NRVSGALEKAIIIYDSPTKTLSVVVTHQNGQ--------ITTVAQQIDLKVVLPKEVS 721
Query: 240 VGFSGATGQL-VEQHRIHVWDFKSSF 264
VGFS T E+H IH W F S+F
Sbjct: 722 VGFSATTWNTHRERHDIHSWSFTSTF 799
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 100 bits (248), Expect = 3e-22
Identities = 55/147 (37%), Positives = 83/147 (56%)
Frame = +3
Query: 6 LFVILISLFMVAHNVNSETFTFPGFQLPNTKDITFEGDAFASNGVLELTKTAYGVPLPSS 65
L ++L++ +S +F F +T I +EGD ++NG ++L K +Y +
Sbjct: 54 LLLLLLTTTTTFPTTHSLSFNITNFD-DDTTAIAYEGDGKSTNGSIDLNKVSYFFRV--- 221
Query: 66 SGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSKIPKNSTG 125
GRA A+P+HL+D T L FTT FT + + +GDG AF++AP +IP NS G
Sbjct: 222 -GRAISAQPLHLYDPTTKTLTDFTTRFTFTISAINKTSYGDGFAFYLAPLGYQIPPNSAG 398
Query: 126 GFLGLFSKDSAFDSYQNQIVAVEFDTY 152
G GLF+ + + QN +VAVEFDT+
Sbjct: 399 GTFGLFNATTNNNLPQNHVVAVEFDTF 479
>TC10090 weakly similar to UP|Q9ZWP6 (Q9ZWP6) Lectin, partial (43%)
Length = 461
Score = 91.7 bits (226), Expect = 1e-19
Identities = 55/136 (40%), Positives = 80/136 (58%), Gaps = 4/136 (2%)
Frame = +3
Query: 3 LMSLFVILISLFMVAHNVNSE---TFTFPGFQLPNTKDITFEGDAF-ASNGVLELTKTAY 58
L++LF +L+ + VNS +FTF F P D+ F+GDA A G L+LTK
Sbjct: 72 LLTLFFLLL-----VNKVNSTDYLSFTFNQFT-PQQPDLLFQGDALVAPTGKLQLTKVEN 233
Query: 59 GVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSSGFHGDGIAFFIAPFNSK 118
+P+ S+GRA + PVH+WD KTG +A+F TSF+ + + DG+AFF+AP +++
Sbjct: 234 DLPVYKSTGRALYVAPVHIWDSKTGNVASFITSFSFIIDSPNVNKIADGLAFFLAPVDTQ 413
Query: 119 IPKNSTGGFLGLFSKD 134
K GG LG+F D
Sbjct: 414 PQK--PGGLLGIFDND 455
>TC17134 weakly similar to UP|LEC_BOWMI (P42088) Lectin (Agglutinin) (BMA),
partial (16%)
Length = 741
Score = 57.4 bits (137), Expect = 2e-09
Identities = 55/208 (26%), Positives = 92/208 (43%), Gaps = 37/208 (17%)
Frame = -2
Query: 6 LFVILISLFMVAHNVNSET---------FTFPGFQLPNTK----DITFEGDAFASN--GV 50
+F +L S+ VNS + F FP F L N D+ G A S+ G
Sbjct: 728 MFNVLASIGTYPVTVNSASDVPINVTKHFYFPDFSLNNNPRLEHDVKLLGSAKLSDQKGA 549
Query: 51 LELTKTAYGVPLPSSSGRASFAEPVHLWDKKTGALAAFTTSFTLDVQPTSS--------- 101
L++ + L +GR ++ P+ L D T A+F T+F + +++
Sbjct: 548 LQIPHESQETDLKHLAGRGLYSFPIRLLDPSTKTPASFETTFAFQLHNSTTSVSGGGSGG 369
Query: 102 -----------GFHGDGIAFFIAPFNSKIPKNSTGGFLGLFSKDSAFDSYQNQIVAVEFD 150
G G G+ F I + +G +LG+ + D+ ++Y+ VAVEFD
Sbjct: 368 GGVGGAAAADVGGGGSGLTFMIV--QDEFTVGRSGPWLGMLN-DACENAYK--AVAVEFD 204
Query: 151 TYA--DTWDPFTSHIGIDINSIISSKTV 176
T + DP +H+G+++ SIIS+K +
Sbjct: 203 TRMSPEFGDPNDNHVGVNLGSIISTKII 120
>TC18976 similar to UP|LCB2_ROBPS (Q42372) Bark agglutinin I, polypeptide B
precursor (RPbAI) (LECRPA2), partial (14%)
Length = 617
Score = 39.7 bits (91), Expect = 5e-04
Identities = 20/39 (51%), Positives = 26/39 (66%), Gaps = 3/39 (7%)
Frame = +1
Query: 227 VVDLRTILPEWVRVGFSGATG---QLVEQHRIHVWDFKS 262
VVDL+ +LPE+VRVGFS TG VE + + W F+S
Sbjct: 4 VVDLKNVLPEYVRVGFSATTGLNPDHVETNDVLSWSFES 120
>BP035176
Length = 444
Score = 38.5 bits (88), Expect = 0.001
Identities = 18/41 (43%), Positives = 25/41 (60%)
Frame = -3
Query: 224 LSFVVDLRTILPEWVRVGFSGATGQLVEQHRIHVWDFKSSF 264
LS +D+ L +++ VGFSG+T E HR+ W F SSF
Sbjct: 367 LSMNLDMGQYLNDFMYVGFSGSTQGSTETHRVEWWSFSSSF 245
>TC12079
Length = 414
Score = 26.9 bits (58), Expect = 3.6
Identities = 25/102 (24%), Positives = 43/102 (41%), Gaps = 20/102 (19%)
Frame = +1
Query: 142 NQIVAVEFDTYADTWDPFTSHIGIDINSIISSKTVPWRTGN-----------SLST---- 186
+Q + ++ +T W P + + S+ + PWR+GN S+ST
Sbjct: 106 SQSIPIDDETTVHYWSPKIENPQKPSLLLXSTDSAPWRSGNGANRTNSSLLTSISTXP*P 285
Query: 187 ----SAAFATVSYEPVTKTLSVLVKQIDQQKGLK-FSTITTS 223
A T S E + + V ++D++ GLK F + TS
Sbjct: 286 XVFXXADPXTTSAERSERFQAATVGKLDEKLGLKRFHVVGTS 411
>BP083453
Length = 382
Score = 25.8 bits (55), Expect = 8.0
Identities = 15/50 (30%), Positives = 23/50 (46%)
Frame = +1
Query: 159 FTSHIGIDINSIISSKTVPWRTGNSLSTSAAFATVSYEPVTKTLSVLVKQ 208
F HI I + +S W +G STS ++ + TK ++VKQ
Sbjct: 31 FICHISQSI*DLTTSHAQNWTSGTWSSTSYSYPNPHHGISTKQSQLVVKQ 180
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.134 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,033,878
Number of Sequences: 28460
Number of extensions: 48029
Number of successful extensions: 227
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 218
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 219
length of query: 266
length of database: 4,897,600
effective HSP length: 89
effective length of query: 177
effective length of database: 2,364,660
effective search space: 418544820
effective search space used: 418544820
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0541.9


