

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0487.13
(394 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
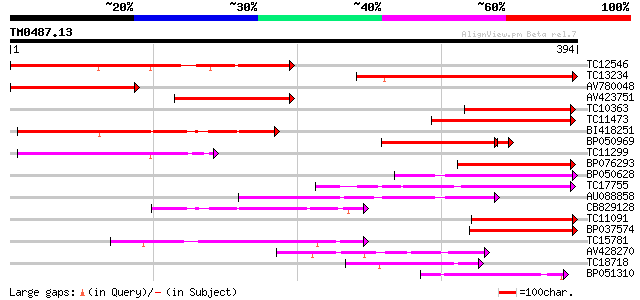
Score E
Sequences producing significant alignments: (bits) Value
TC12546 weakly similar to UP|Q9FJ30 (Q9FJ30) Similarity to heat ... 204 2e-53
TC13234 202 6e-53
AV780048 179 7e-46
AV423751 169 7e-43
TC10363 160 3e-40
TC11473 145 1e-35
BI418251 137 2e-33
BP050969 115 1e-26
TC11299 114 4e-26
BP076293 104 3e-23
BP050628 95 2e-20
TC17755 94 3e-20
AU088858 92 1e-19
CB829128 92 1e-19
TC11091 89 1e-18
BP037574 88 2e-18
TC15781 88 3e-18
AV428270 86 1e-17
TC18718 79 2e-15
BP051310 73 7e-14
>TC12546 weakly similar to UP|Q9FJ30 (Q9FJ30) Similarity to heat shock
transcription factor HSF30, partial (9%)
Length = 755
Score = 204 bits (519), Expect = 2e-53
Identities = 123/217 (56%), Positives = 149/217 (67%), Gaps = 19/217 (8%)
Frame = +2
Query: 1 MKKMKMEDRFSSLPDPILCHILSLLTTKEAVTTSILSKRWIPLWRSVPTLDFDDSNLYGG 60
MKKMKMED S+LPD +LCHILS LT+KEAV TS+LS+RWIPLWRSVPTL F D+N +
Sbjct: 128 MKKMKMEDGISTLPDALLCHILSFLTSKEAVATSVLSRRWIPLWRSVPTLHFKDANYHTD 307
Query: 61 ---------DEVYARFVEAVYKVILARDFNQPIKKFRLVLTERLK---DPANISVWVNLV 108
+V +R V++VY VIL+RDF PIKKF L L + + DPAN+SVWVN V
Sbjct: 308 IGHADHDIVKDVRSRHVQSVYAVILSRDFQLPIKKFYLRLNDVCQPFYDPANVSVWVNAV 487
Query: 109 LKRRVEHIDISLHFEDDEFLELPHIVVDTP-----SMFSCTTLVVLKLE-GLELKANFSS 162
++R++EH+DIS LP+ ++ TP S+FSC TLVVLKL GLELK F S
Sbjct: 488 VQRQLEHLDIS----------LPYPMLSTPRANLSSIFSCRTLVVLKLRGGLELK-RFPS 634
Query: 163 VDLPFLKVLHLQD-LLLQDEGCLAEILSGCLALEDLK 198
V P LKVLHLQ LLL D LAE+LSGC LE+LK
Sbjct: 635 VHFPCLKVLHLQGALLLHDVPYLAELLSGCPVLENLK 745
>TC13234
Length = 609
Score = 202 bits (515), Expect = 6e-53
Identities = 102/157 (64%), Positives = 120/157 (75%), Gaps = 4/157 (2%)
Frame = +3
Query: 242 SFLRIHEIDYNLI-YMGED--MFHNLTHLELVYTTFNRDWFEVLEFLKYCPKLEVLVIKQ 298
+FLRI +ID+ L+ Y GED MFHNLT +ELVYT RDW +V+EFLKYCPKL+VLVIKQ
Sbjct: 3 NFLRIRDIDFRLLPYGGEDIDMFHNLTRVELVYTGHTRDWLDVVEFLKYCPKLQVLVIKQ 182
Query: 299 PQFYNVY-LNKLGAKDWQYPSSVPECILLHLKECCLNHYRGTKGELQFAKYIMEHGRLLN 357
PQ Y + +L DWQYPSSVP+CIL HLKEC LN+YRGTKGE +FA+YIM +GR L
Sbjct: 183 PQVDGYYTVGELRGNDWQYPSSVPKCILFHLKECYLNNYRGTKGEFKFARYIMRNGRFLK 362
Query: 358 KMTICSSTAEKQGEKLENLKKLFSCTRCSATCKFSFK 394
KMTIC TA KQ K ++KKLFSC+ SATCK FK
Sbjct: 363 KMTICKCTAVKQLGKPNDIKKLFSCSARSATCKLYFK 473
>AV780048
Length = 501
Score = 179 bits (454), Expect = 7e-46
Identities = 88/90 (97%), Positives = 88/90 (97%)
Frame = +2
Query: 1 MKKMKMEDRFSSLPDPILCHILSLLTTKEAVTTSILSKRWIPLWRSVPTLDFDDSNLYGG 60
MKKMKMEDRFSSLPDPILCHILSLLTTKEAVTTSILSKRWIPLWRSVPTLDFDDSN YGG
Sbjct: 230 MKKMKMEDRFSSLPDPILCHILSLLTTKEAVTTSILSKRWIPLWRSVPTLDFDDSNWYGG 409
Query: 61 DEVYARFVEAVYKVILARDFNQPIKKFRLV 90
DEVYAR VEAVYKVILARDFNQPIKKFRLV
Sbjct: 410 DEVYARLVEAVYKVILARDFNQPIKKFRLV 499
>AV423751
Length = 254
Score = 169 bits (428), Expect = 7e-43
Identities = 84/84 (100%), Positives = 84/84 (100%)
Frame = +2
Query: 115 HIDISLHFEDDEFLELPHIVVDTPSMFSCTTLVVLKLEGLELKANFSSVDLPFLKVLHLQ 174
HIDISLHFEDDEFLELPHIVVDTPSMFSCTTLVVLKLEGLELKANFSSVDLPFLKVLHLQ
Sbjct: 2 HIDISLHFEDDEFLELPHIVVDTPSMFSCTTLVVLKLEGLELKANFSSVDLPFLKVLHLQ 181
Query: 175 DLLLQDEGCLAEILSGCLALEDLK 198
DLLLQDEGCLAEILSGCLALEDLK
Sbjct: 182 DLLLQDEGCLAEILSGCLALEDLK 253
>TC10363
Length = 387
Score = 160 bits (405), Expect = 3e-40
Identities = 75/77 (97%), Positives = 75/77 (97%)
Frame = +3
Query: 317 PSSVPECILLHLKECCLNHYRGTKGELQFAKYIMEHGRLLNKMTICSSTAEKQGEKLENL 376
PSSVPECILLHLKECCLNHYRGTKGELQFAKYIMEHGRLLNKMTICSS AEKQGEKLENL
Sbjct: 3 PSSVPECILLHLKECCLNHYRGTKGELQFAKYIMEHGRLLNKMTICSSPAEKQGEKLENL 182
Query: 377 KKLFSCTRCSATCKFSF 393
KKLFSC RCSATCKFSF
Sbjct: 183 KKLFSCPRCSATCKFSF 233
>TC11473
Length = 435
Score = 145 bits (365), Expect = 1e-35
Identities = 71/100 (71%), Positives = 79/100 (79%)
Frame = +2
Query: 294 LVIKQPQFYNVYLNKLGAKDWQYPSSVPECILLHLKECCLNHYRGTKGELQFAKYIMEHG 353
LVI QP FY+ L++L DWQ P SVP+CILLHLK C LN YRGTKGELQFA+YIM HG
Sbjct: 2 LVINQPDFYDYDLDQLHISDWQDPPSVPKCILLHLKVCYLNDYRGTKGELQFARYIMRHG 181
Query: 354 RLLNKMTICSSTAEKQGEKLENLKKLFSCTRCSATCKFSF 393
R L +MTI SS AE Q KL+N+KKLFSCTR SATCKFSF
Sbjct: 182 RFLKRMTISSSPAENQRGKLKNVKKLFSCTRRSATCKFSF 301
>BI418251
Length = 546
Score = 137 bits (346), Expect = 2e-33
Identities = 83/185 (44%), Positives = 114/185 (60%), Gaps = 3/185 (1%)
Frame = +1
Query: 6 MEDRFSSLPDPILCHILSLLTTKEAVTTSILSKRWIPLWRSVPTLDFDDSNLYGGD--EV 63
M DR S LPD +LC ILS L T++AV TS+L KRW LW SVPTLDFDD G ++
Sbjct: 31 MADRISKLPDEVLCQILSFLPTEDAVATSVLCKRWSSLWLSVPTLDFDDYRYLKGKKLKL 210
Query: 64 YARFVEAVYKVILARDFNQPIKKFRL-VLTERLKDPANISVWVNLVLKRRVEHIDISLHF 122
+ F+ VY +IL+R +QPIK F L V++E P ++ VW+N ++R+VE++DI L
Sbjct: 211 QSSFINFVYAIILSRALHQPIKNFTLSVISEECPYP-DVKVWLNAAMQRQVENLDICLSL 387
Query: 123 EDDEFLELPHIVVDTPSMFSCTTLVVLKLEGLELKANFSSVDLPFLKVLHLQDLLLQDEG 182
LP S+ SCTTLVVLKL ++ FS VDLP LK LHL+ +++ +
Sbjct: 388 S-----TLP------CSILSCTTLVVLKLSDVKFHV-FSCVDLPSLKTLHLELVVILNPQ 531
Query: 183 CLAEI 187
L ++
Sbjct: 532 SLMDL 546
>BP050969
Length = 557
Score = 115 bits (288), Expect(2) = 1e-26
Identities = 52/82 (63%), Positives = 61/82 (73%)
Frame = -3
Query: 259 DMFHNLTHLELVYTTFNRDWFEVLEFLKYCPKLEVLVIKQPQFYNVYLNKLGAKDWQYPS 318
DMFHNLTH+E+VY FN DWFEV+EFL +CPKL+ LVI QP+F+ L + QYP
Sbjct: 306 DMFHNLTHVEIVYANFNEDWFEVVEFLTHCPKLQTLVISQPKFHEYGLGEYEVGFGQYPL 127
Query: 319 SVPECILLHLKECCLNHYRGTK 340
VP+CIL HLKEC LN YRGTK
Sbjct: 126 HVPKCILRHLKECYLNDYRGTK 61
Score = 37.7 bits (86), Expect = 0.003
Identities = 20/38 (52%), Positives = 28/38 (73%)
Frame = -1
Query: 212 EFITLPKLVRADISESGGSQHFMMKVVNNVSFLRIHEI 249
+F TLPKLVRA ISE + +++V+NNV FLRI ++
Sbjct: 539 KFKTLPKLVRAHISEG----YPIVEVINNVKFLRIDQV 438
Score = 21.2 bits (43), Expect(2) = 1e-26
Identities = 8/11 (72%), Positives = 9/11 (81%)
Frame = -2
Query: 340 KGELQFAKYIM 350
KGE FA+YIM
Sbjct: 64 KGEFXFARYIM 32
>TC11299
Length = 498
Score = 114 bits (284), Expect = 4e-26
Identities = 68/145 (46%), Positives = 88/145 (59%), Gaps = 5/145 (3%)
Frame = +1
Query: 6 MEDRFSSLPDPILCHILSLLTTKEAVTTSILSKRWIPLWRSVPTLDFDDSNLYGGDEVYA 65
M DR S+LPD I+CHILS L T+ AV T++LSKRW LWRSV LDF +Y D+
Sbjct: 79 MVDRISALPDEIICHILSFLPTENAVATAVLSKRWTHLWRSVSALDFSSVRMYEPDDHRF 258
Query: 66 RFVEAVYKVILARDFNQPIKKFRLVLTERLK----DPANISVWVNLVLKRRVEHID-ISL 120
F + VY V+L R+ PIKKF L L + + D ANI WVN V + RV I+ + L
Sbjct: 259 FFSDIVYSVLLFRNAATPIKKFSLELDDDVAVANLDIANIHKWVNFVTQCRVGGIEHLRL 438
Query: 121 HFEDDEFLELPHIVVDTPSMFSCTT 145
HF D F++LP + + S+ SC T
Sbjct: 439 HFMD--FIQLPQLPI---SILSCKT 498
>BP076293
Length = 485
Score = 104 bits (259), Expect = 3e-23
Identities = 50/82 (60%), Positives = 58/82 (69%)
Frame = -2
Query: 312 KDWQYPSSVPECILLHLKECCLNHYRGTKGELQFAKYIMEHGRLLNKMTICSSTAEKQGE 371
+DWQ PECI LHLK C LN YRGT+GE QFA+YI+++GR+L M ICS KQ E
Sbjct: 469 RDWQCSLPGPECIQLHLKRCYLNDYRGTEGEFQFARYILQNGRVLESMIICSFPDVKQQE 290
Query: 372 KLENLKKLFSCTRCSATCKFSF 393
K ENLKKL SCTR S TC+ F
Sbjct: 289 KHENLKKLSSCTRLSPTCELLF 224
>BP050628
Length = 523
Score = 94.7 bits (234), Expect = 2e-20
Identities = 48/127 (37%), Positives = 74/127 (57%)
Frame = -3
Query: 268 ELVYTTFNRDWFEVLEFLKYCPKLEVLVIKQPQFYNVYLNKLGAKDWQYPSSVPECILLH 327
EL+ + +W VL LK CPKL+ L++ ++ + W P + PEC+
Sbjct: 521 ELMMSGIGMNWHLVLAMLKKCPKLQKLLL------DMKASSTHDMVWTSPFNDPECLSSQ 360
Query: 328 LKECCLNHYRGTKGELQFAKYIMEHGRLLNKMTICSSTAEKQGEKLENLKKLFSCTRCSA 387
L++C + +Y+GTK EL FA YIM+ R+L M I ++ + +QG+K E +K+L C R SA
Sbjct: 359 LRKCSITNYKGTKSELHFATYIMQSSRVLQTMAIDTAPSSEQGDKFEMIKELSLCPRSSA 180
Query: 388 TCKFSFK 394
C+ SFK
Sbjct: 179 ICQLSFK 159
>TC17755
Length = 776
Score = 94.4 bits (233), Expect = 3e-20
Identities = 67/182 (36%), Positives = 92/182 (49%), Gaps = 1/182 (0%)
Frame = +2
Query: 213 FITLPKLVRADISESGGSQHFMMKVVNNVSFLRIHEIDYNLIYMGEDMFHNLTHLELVYT 272
F +L KLVRADI G F+ E D L + +F NLTH EL+
Sbjct: 74 FKSLSKLVRADIHAIGFD-----------IFVESPEFDKYLDDI--PLFPNLTHAELMLG 214
Query: 273 TFNRDWFEVLEFLKYCPKLEVLVIKQPQFYNVYLNKLGAKDWQYPSSVPECILLHLKECC 332
F+ W VL LK+CP L+ +V+ Y Y + W P VPEC+ L+ C
Sbjct: 215 -FDMSWDLVLAILKHCPMLQNVVLDMQTHYAYYKEPV----WISPCCVPECLSSQLRTCD 379
Query: 333 LNHYRGTKGELQFAKYIMEHGRLLNKMTICS-STAEKQGEKLENLKKLFSCTRCSATCKF 391
+ + + EL FAKYIM++ R+L MTI + +E +K+E LK+LF C R S C+
Sbjct: 380 IINSEVKESELHFAKYIMQNSRVLRTMTIWTRGLSELNVDKIELLKELFLCPRSSTICEL 559
Query: 392 SF 393
SF
Sbjct: 560 SF 565
>AU088858
Length = 765
Score = 92.0 bits (227), Expect = 1e-19
Identities = 65/183 (35%), Positives = 96/183 (51%), Gaps = 2/183 (1%)
Frame = +3
Query: 160 FSSVDLPFLKVLHLQDLLLQDEGCLAEILSGCLALEDLK-ARDVFFD-GNKADAEFITLP 217
FS V LP LK L L L+ + + E+LSGC LE + +R + D + ++ +L
Sbjct: 6 FSFVHLPSLKTLQLYCLIFPEPQYVMELLSGCPILEYMHLSRMRYADCSSPSNTNVKSLT 185
Query: 218 KLVRADISESGGSQHFMMKVVNNVSFLRIHEIDYNLIYMGEDMFHNLTHLELVYTTFNRD 277
KLV ADIS G +KV+ NV FLRI + Y + +F NLTHLEL+ + +
Sbjct: 186 KLVSADISFIGFE--IPLKVICNVEFLRIDKYPYEI-----PVFPNLTHLELMMSGIGMN 344
Query: 278 WFEVLEFLKYCPKLEVLVIKQPQFYNVYLNKLGAKDWQYPSSVPECILLHLKECCLNHYR 337
W VL LK CPKL+ L++ ++ + W P + P C+ L +C + +Y+
Sbjct: 345 WHLVLAMLKKCPKLQKLLL------DMKASSTHDMVWTSPFNDPXCLSSQLXKCSITNYK 506
Query: 338 GTK 340
GTK
Sbjct: 507 GTK 515
>CB829128
Length = 463
Score = 92.0 bits (227), Expect = 1e-19
Identities = 62/153 (40%), Positives = 91/153 (58%), Gaps = 2/153 (1%)
Frame = +3
Query: 99 ANISVWVNLVLKRRVEHIDISLHFEDDEFLELPHIVVDTPSMFSCTTLVVLKLEGLELKA 158
+++ VW+N ++R+VE+++I L ++P S+ CTTLV+LKL+ + A
Sbjct: 18 SDLKVWLNAAIQRQVENLEIELTDS-----QMPC------SILRCTTLVILKLDSVTFDA 164
Query: 159 NFSSVDLPFLKVLHLQDLLLQDEGCLAEILSGCLALEDLKARDVFFDGNKADAEFITLPK 218
F+SV LP LK LHL ++ L CL ++L GC LEDLK + F DG+ + + TL K
Sbjct: 165 -FTSVYLPSLKTLHLFEVDLVKAQCLIDLLYGCPILEDLKTQIYFRDGS-FEGKVKTLSK 338
Query: 219 LVRADISESGGSQHFM--MKVVNNVSFLRIHEI 249
LVRAD+ G HF+ +K NV FLRI E+
Sbjct: 339 LVRADVFLDG---HFIIPVKAFQNVEFLRIEEV 428
>TC11091
Length = 502
Score = 89.0 bits (219), Expect = 1e-18
Identities = 42/73 (57%), Positives = 54/73 (73%)
Frame = +3
Query: 322 ECILLHLKECCLNHYRGTKGELQFAKYIMEHGRLLNKMTICSSTAEKQGEKLENLKKLFS 381
ECILLHL+EC LN YR T+GE QFA+++M++GR L KMT CS Q +LE ++KL S
Sbjct: 3 ECILLHLEECYLNDYRXTQGEFQFARHVMQNGRSLKKMTTCSGDFINQQGRLELIEKLSS 182
Query: 382 CTRCSATCKFSFK 394
CTR SATC+ F+
Sbjct: 183 CTRHSATCELYFE 221
>BP037574
Length = 431
Score = 88.2 bits (217), Expect = 2e-18
Identities = 44/75 (58%), Positives = 51/75 (67%)
Frame = -2
Query: 320 VPECILLHLKECCLNHYRGTKGELQFAKYIMEHGRLLNKMTICSSTAEKQGEKLENLKKL 379
VPE ILL LK C LN YRGT+ E QFA+YIM++G L M IC S Q KL+NL++L
Sbjct: 430 VPEYILLRLKRCYLNEYRGTRAEFQFARYIMQNGIFLENMAICCSYEVNQEGKLKNLQEL 251
Query: 380 FSCTRCSATCKFSFK 394
CTR SA CK SFK
Sbjct: 250 SLCTRKSAACKLSFK 206
>TC15781
Length = 621
Score = 87.8 bits (216), Expect = 3e-18
Identities = 67/184 (36%), Positives = 98/184 (52%), Gaps = 5/184 (2%)
Frame = +2
Query: 71 VYKVILARDFNQPIKKFRLVL--TERLKDPANISVWVNLVLKRRVEHIDISLHFEDDEFL 128
V ILARD QPI +F L+ T L+ + W+N V+ RR+E+++I +
Sbjct: 26 VNATILARDERQPITRF*LLYESTVSLEGSDLVHDWLNTVMPRRIENLEICIP------- 184
Query: 129 ELPHIVVDTPSMFSCTTLVVLKLEGLELKANFSS-VDLPFLKVLHLQDLLLQDEGCLAEI 187
P ++SC TLVVLKL+G L SS V+LP LK LHL+ + D ++
Sbjct: 185 --PLSRGIGLKIYSCRTLVVLKLKGYPLGVYVSSHVELPSLKSLHLEKVHFVDFQSFLKL 358
Query: 188 LSGCLALEDLKARDVFFDGNKADAE--FITLPKLVRADISESGGSQHFMMKVVNNVSFLR 245
+ GC LEDL+A V + D E F +LPKLV I++ + F +K+++N FL
Sbjct: 359 ICGCPLLEDLEAGTVSYLDTSFDQEFRFRSLPKLVSVVINDY--NFRFPLKIISNAEFLS 532
Query: 246 IHEI 249
I E+
Sbjct: 533 IEEV 544
>AV428270
Length = 428
Score = 85.5 bits (210), Expect = 1e-17
Identities = 59/155 (38%), Positives = 84/155 (54%), Gaps = 7/155 (4%)
Frame = +1
Query: 186 EILSGCLALEDLKARDVFFDGNKA-----DAEFITLPKLVRADISESGGSQHFMMKVVNN 240
E L+GC LE+LKAR ++ G + + + LPKL+RADI S + M++ N
Sbjct: 1 EFLAGCPILENLKARRIWITGESSFYRGGELADLGLPKLIRADIVRS---RLPMLQPFYN 171
Query: 241 VSFLR--IHEIDYNLIYMGEDMFHNLTHLELVYTTFNRDWFEVLEFLKYCPKLEVLVIKQ 298
V+FLR I E+ Y Y FHNLTHLELV +N ++E LK+CP L+ LV ++
Sbjct: 172 VAFLRVDIEEVHYAFPY-----FHNLTHLELVIELYNS--LVLVEMLKHCPNLQSLVFER 330
Query: 299 PQFYNVYLNKLGAKDWQYPSSVPECILLHLKECCL 333
+ ++W P VPEC+ +LK C L
Sbjct: 331 LD----EAQRCDPENWIDPEFVPECLSFNLKTCTL 423
>TC18718
Length = 518
Score = 78.6 bits (192), Expect = 2e-15
Identities = 46/104 (44%), Positives = 60/104 (57%), Gaps = 8/104 (7%)
Frame = +1
Query: 234 MMKVVNNVSFLRIHEIDYNLIY--------MGEDMFHNLTHLELVYTTFNRDWFEVLEFL 285
+M+ V NV FLRI+E+ + G FHNLTH+EL Y DW V+E L
Sbjct: 7 LMEAVKNVEFLRIYEMQRLRRHDDGEGAGPYGFPKFHNLTHIELEYNCRISDWLTVVELL 186
Query: 286 KYCPKLEVLVIKQPQFYNVYLNKLGAKDWQYPSSVPECILLHLK 329
K+CPKL+VLVI Q + + + +DW+YP SVPE I L LK
Sbjct: 187 KHCPKLQVLVINQSCLPDDFGEVV--EDWEYPPSVPESIQLCLK 312
>BP051310
Length = 467
Score = 73.2 bits (178), Expect = 7e-14
Identities = 39/108 (36%), Positives = 61/108 (56%), Gaps = 5/108 (4%)
Frame = -3
Query: 286 KYCPKLEVLVIKQPQFYNVYLNKLGAKDWQYPSSVPECILLHLKECCLNHYRGTKGELQF 345
K+CPKL+ LV+++P+ + L +W P VPEC+ L+LK C ++G K E Q
Sbjct: 465 KHCPKLQSLVLERPE-ESRDLENCDLGNWVDPDFVPECLALNLKSCTFRDFKGLKVEFQL 289
Query: 346 AKYIMEHGRLLNKMTICSSTAEKQGEKLENLKKLFSC-----TRCSAT 388
A Y++++ R+L MTIC+ + Q +KKL+S +RC T
Sbjct: 288 ASYVLKNARVLETMTICNQQSISQ----RQIKKLYSAALRSPSRCQIT 157
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,535,152
Number of Sequences: 28460
Number of extensions: 108070
Number of successful extensions: 842
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 808
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 816
length of query: 394
length of database: 4,897,600
effective HSP length: 92
effective length of query: 302
effective length of database: 2,279,280
effective search space: 688342560
effective search space used: 688342560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0487.13


