

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
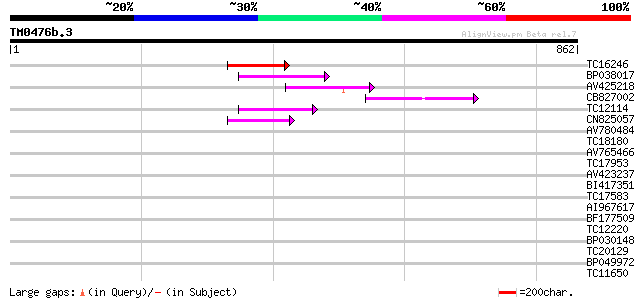
Query= TM0476b.3
(862 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired re... 103 9e-23
BP038017 100 8e-22
AV425218 91 8e-19
CB827002 76 3e-14
TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transp... 69 4e-12
CN825057 50 2e-06
AV780484 38 0.008
TC18180 weakly similar to GB|AAF04891.1|6175165|ATAC011437 Mutat... 37 0.013
AV765466 36 0.023
TC17953 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F17... 36 0.023
AV423237 36 0.030
BI417351 35 0.066
TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired re... 34 0.11
AI967617 33 0.19
BF177509 33 0.25
TC12220 32 0.56
BP030148 30 2.1
TC20129 homologue to GB|AAF04891.1|6175165|ATAC011437 Mutator-li... 29 2.8
BP049972 29 2.8
TC11650 similar to GB|AAP47462.1|32331869|AY164888 RTNLB1 {Arabi... 29 3.6
>TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired response
protein-like, partial (4%)
Length = 660
Score = 103 bits (258), Expect = 9e-23
Identities = 45/97 (46%), Positives = 65/97 (66%), Gaps = 2/97 (2%)
Frame = +2
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVSDG--RSALDYLASLGTKDPSMFVRHTEDADKNLEH 388
G+ + + DL N + K +R + +G +AL YL DP F + T+ D++LE+
Sbjct: 68 GHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKADNDPMFFYKFTKTGDESLEN 247
Query: 389 LFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIF 425
LFWCDG+ RM+Y+VFGD +AFD+TYKKNKY +PLV+F
Sbjct: 248 LFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVF 358
>BP038017
Length = 570
Score = 100 bits (250), Expect = 8e-22
Identities = 46/138 (33%), Positives = 78/138 (56%)
Frame = +2
Query: 349 QRRAHVSDGRSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALA 408
Q+R D ++ L+Y + K+P + D + + ++FW D R Y+ FGDA+
Sbjct: 146 QKRTLGRDAQNLLNYFKKMQGKNPGFYYAIQLDDENRMINVFWADARSRSAYNYFGDAVI 325
Query: 409 FDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTT 468
FD Y+ N+Y+ P F+ VNH Q + G A++ +E E ++ WL L AM+ + P +
Sbjct: 326 FDTMYRPNQYQVPFAPFTGVNHHGQNVLFGCALLLDESESSFTWLFRTWLSAMNDRPPVS 505
Query: 469 VITDGDKAMRNAIRRVFP 486
+ TD D+A++ A+ +VFP
Sbjct: 506 ITTDQDRAIQAAVAQVFP 559
>AV425218
Length = 419
Score = 90.9 bits (224), Expect = 8e-19
Identities = 43/139 (30%), Positives = 72/139 (50%), Gaps = 4/139 (2%)
Frame = +2
Query: 420 RPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKSPTTVITDGDKAMRN 479
RPL VNH Q+ + G A++ +E E++VWL + LL M P +ITD +AM+
Sbjct: 2 RPLATLVGVNHHGQSVLFGCALLSSEDSESFVWLFQSLLHCMSGVPPQGIITDHSEAMKK 181
Query: 480 AIRRVFPNASHRLCAWHLMCNATDNI----KIPDFLPKFKTCMFGDFQIGDFKRKWESLV 535
AI V P+ HR C ++M + + + ++ I +F+R W+ +V
Sbjct: 182 AIETVLPSTRHRWCLSYIMKKLPQKLLGYAQYESIRHHLQNVVYDAVVIDEFERNWKKIV 361
Query: 536 EEFNLHENPWIIDMYARRH 554
E+F L +N W+ +++ RH
Sbjct: 362 EDFGLEDNEWLNELFLERH 418
>CB827002
Length = 534
Score = 75.9 bits (185), Expect = 3e-14
Identities = 45/172 (26%), Positives = 77/172 (44%)
Frame = +1
Query: 542 ENPWIIDMYARRHTWAAAYFRGKFFAGFRTTSRCEGFNAQLGKFVKRRYNLKEFLQHFHH 601
++ W+ +Y+ A + R FFA T R + N+ +V NL +F + +
Sbjct: 31 DHEWL-SLYSSCRQGAPVHLRDTFFAEMSITQRSDSMNSYFDGYVNASTNLNQFFKLYEK 207
Query: 602 CLEYMRHREVEADFRSGYGDPGLKAPPIFQSIELSASRILTRQLFFRFRPTIVSAADMII 661
LE +EV AD+ + P L+ P +E AS + TR++F RF+ +V +
Sbjct: 208 ALESRNEKEVRADYDTMNTLPVLRTP---SPMEKQASELYTRKIFMRFQEELVGTLTFMA 378
Query: 662 TNCEDTLGMSMHTVKRSQESSREWLVSYYTTTYEFRCSCMRMESIGLACAHI 713
+ +D + + V + E + + V + + CSC E GL C HI
Sbjct: 379 SKADDDGEVITYHVAKFGEDHKAYYVKFNVLEMKASCSCQMFEFSGLLCRHI 534
>TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transposase-like
{Oryza sativa (japonica cultivar-group);}, partial (8%)
Length = 488
Score = 68.6 bits (166), Expect = 4e-12
Identities = 36/122 (29%), Positives = 62/122 (50%), Gaps = 2/122 (1%)
Frame = +1
Query: 348 KQRRAHVSDGRSA--LDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGD 405
K+R+ + G + L Y ++ + + D + + ++FW D ++Y FGD
Sbjct: 13 KRRQRSLLHGEAGYILQYFQRKLVENSPFYHAYQLDDEDQITNVFWVDARMLIDYGYFGD 192
Query: 406 ALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVIGNEKEETYVWLLEQLLEAMDKKS 465
++ D+TY + RPL +FS NH + + G A++ +E E+Y WL E LEA +S
Sbjct: 193 MVSLDSTYCTHSSNRPLAVFSGFNHHRKAVIFGAALLYDETTESY*WLFESFLEAHKHES 372
Query: 466 PT 467
T
Sbjct: 373 IT 378
>CN825057
Length = 721
Score = 50.1 bits (118), Expect = 2e-06
Identities = 28/105 (26%), Positives = 52/105 (48%), Gaps = 2/105 (1%)
Frame = +1
Query: 331 GYNNTQYILKDLQNQVGKQRRAHVSDGRSA--LDYLASLGTKDPSMFVRHTEDADKNLEH 388
G N + ++ DL +Q K + + +G + L+Y + ++P+ F + ++ L +
Sbjct: 397 GCLNIESLVGDLNDQFKKGQYLAMDEGDAQVMLEYFKHIQKENPNFFYSIDLNEEQRLRN 576
Query: 389 LFWCDGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQ 433
+FW D +Y F D ++FD +Y K+ + P F VNH Q
Sbjct: 577 IFWIDAKSINDYLSFNDVVSFDTSYIKSNEKLPFAPFVGVNHHCQ 711
>AV780484
Length = 529
Score = 37.7 bits (86), Expect = 0.008
Identities = 18/64 (28%), Positives = 32/64 (49%)
Frame = -1
Query: 358 RSALDYLASLGTKDPSMFVRHTEDADKNLEHLFWCDGIGRMNYSVFGDALAFDATYKKNK 417
++ +Y S ++P+ F D +++L +FW D GR++Y + Y KNK
Sbjct: 205 KAMTEYFVSTQGENPNFFYAIDLDLNRHLTSVFWVDIKGRLDYETSMMLVLIHTHYLKNK 26
Query: 418 YRRP 421
Y+ P
Sbjct: 25 YKIP 14
>TC18180 weakly similar to GB|AAF04891.1|6175165|ATAC011437 Mutator-like
transposase {Arabidopsis thaliana;} , partial (4%)
Length = 513
Score = 37.0 bits (84), Expect = 0.013
Identities = 30/100 (30%), Positives = 41/100 (41%), Gaps = 6/100 (6%)
Frame = +3
Query: 764 CMLEACRKMCTLACPLIDDFRQTWEIVGGHTQHLENKNNP----SVPEGVTGTPSANAGL 819
C+ + C + CT + +R T+ + NP +VPE P
Sbjct: 3 CVYDYCSRYCTT-----ESYRLTYS----------ERVNPIPIMAVPESKDSQPVVTVTP 137
Query: 820 T-THRRRGRPCGPS-TSRLRAKRPLKCSTCKVIGHNRLTC 857
T R GRP S+ KR L CS CK +GHN+ TC
Sbjct: 138 PFTRRPPGRPATKRYVSQNIVKRELHCSRCKGLGHNKSTC 257
>AV765466
Length = 599
Score = 36.2 bits (82), Expect = 0.023
Identities = 13/43 (30%), Positives = 22/43 (50%)
Frame = -2
Query: 697 RCSCMRMESIGLACAHIIAVLVDLNIHNIPKSLVLERWSQGAK 739
+C C E G+ C H +A+L + +P VL+RW + +
Sbjct: 595 KCDCRLFEFRGILCRHSLAILSQERVKEVPDRYVLDRWRKNMR 467
>TC17953 similar to GB|AAM26637.1|20855902|AY101516 AT3g06940/F17A9_9
{Arabidopsis thaliana;}, partial (6%)
Length = 527
Score = 36.2 bits (82), Expect = 0.023
Identities = 25/60 (41%), Positives = 31/60 (51%), Gaps = 1/60 (1%)
Frame = +3
Query: 799 NKNNPSVPEGVTGTPSANAGLTTHRRRGRP-CGPSTSRLRAKRPLKCSTCKVIGHNRLTC 857
N+ ++P VT TP T R GRP P S KR L+CS CK +GHN+ TC
Sbjct: 3 NEGELALP-AVTVTPPP-----TKRPPGRPKMKPVESIDMIKRQLQCSKCKELGHNKKTC 164
>AV423237
Length = 310
Score = 35.8 bits (81), Expect = 0.030
Identities = 25/63 (39%), Positives = 31/63 (48%), Gaps = 2/63 (3%)
Frame = +2
Query: 393 DGIGRMNYSVFGDALAFDATYKKNKYRRPLVIFSCVNHRNQTTVVGG--AVIGNEKEETY 450
D RMNYS FGDA+ FD T + I S + T V A+I NE E ++
Sbjct: 17 DATSRMNYSYFGDAVIFDTTIDNHIES----ICSSWG*SSWATCVIWLVALIANESESSF 184
Query: 451 VWL 453
VWL
Sbjct: 185 VWL 193
>BI417351
Length = 425
Score = 34.7 bits (78), Expect = 0.066
Identities = 20/50 (40%), Positives = 26/50 (52%)
Frame = +2
Query: 791 GGHTQHLENKNNPSVPEGVTGTPSANAGLTTHRRRGRPCGPSTSRLRAKR 840
G H Q ++N PEG P ++A L RR+ RP G RLRA+R
Sbjct: 254 GPHNQQGIGRSNS--PEGAREDPPSHAILPPRRRQARPAGAMHRRLRARR 397
>TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein, mutator-like transposase-like protein,
phytochrome A signaling protein-like, partial (3%)
Length = 666
Score = 33.9 bits (76), Expect = 0.11
Identities = 13/30 (43%), Positives = 17/30 (56%)
Frame = +3
Query: 398 MNYSVFGDALAFDATYKKNKYRRPLVIFSC 427
M+Y FGD ++ D TY N RPL + C
Sbjct: 12 MDYEYFGDVVSLDTTYSTNNAYRPLAVSMC 101
>AI967617
Length = 442
Score = 33.1 bits (74), Expect = 0.19
Identities = 13/19 (68%), Positives = 14/19 (73%)
Frame = +1
Query: 839 KRPLKCSTCKVIGHNRLTC 857
KR L CS CK GHN+LTC
Sbjct: 301 KRVLNCSVCKQPGHNKLTC 357
>BF177509
Length = 472
Score = 32.7 bits (73), Expect = 0.25
Identities = 27/112 (24%), Positives = 50/112 (44%), Gaps = 10/112 (8%)
Frame = +1
Query: 628 PIFQSIELSASRILTRQLFFRFRPTIVSAADMIITNCEDTLGMSM-----HTVKRSQESS 682
PI Q +E +++T F R T + A +++ + E + ++ + V R+ E+
Sbjct: 142 PIIQMMECIRRQLMT--WFNERRETSMQWASILVPSAERCVAEALERARTYQVLRANEAE 315
Query: 683 REWLVSYYTTTYEFR---CSCMRMESIGLACAHIIAVLVDL--NIHNIPKSL 729
E + + + R C C + GL CAH +A L+ N+H + L
Sbjct: 316 FEVISHEGSNIVDIRNRCCLCRGWQLYGLPCAHAVAALLSCRQNVHRFTEKL 471
>TC12220
Length = 473
Score = 31.6 bits (70), Expect = 0.56
Identities = 11/34 (32%), Positives = 19/34 (55%)
Frame = +1
Query: 707 GLACAHIIAVLVDLNIHNIPKSLVLERWSQGAKD 740
G+ C H + V L + +P +L RW++ A+D
Sbjct: 10 GVLCRHPLRVFXILELREVPSRYILHRWTRNAED 111
>BP030148
Length = 398
Score = 29.6 bits (65), Expect = 2.1
Identities = 27/107 (25%), Positives = 42/107 (39%), Gaps = 8/107 (7%)
Frame = +3
Query: 468 TVITDGDKAMRNAIRRVFPNASHRLCAWHLMCNATDNIKIPDFLPKFKTCMFGDF----- 522
T+++D K + + + FP A H C HL F +F M +
Sbjct: 24 TILSDRQKGIVDGVEANFPTAFHGFCMRHLS---------DSFRKEFNNTMLVNLLWEAA 176
Query: 523 ---QIGDFKRKWESLVEEFNLHENPWIIDMYARRHTWAAAYFRGKFF 566
+ +F+ K +EE + WI + R WA AYF G+ F
Sbjct: 177 HVLTVIEFESKILE-IEEISQDAAYWIRRIPPR--LWATAYFEGQRF 308
>TC20129 homologue to GB|AAF04891.1|6175165|ATAC011437 Mutator-like
transposase {Arabidopsis thaliana;} , partial (12%)
Length = 588
Score = 29.3 bits (64), Expect = 2.8
Identities = 21/58 (36%), Positives = 25/58 (42%), Gaps = 7/58 (12%)
Frame = +3
Query: 807 EGVTGTPSANAGLTTHRRRGRPCG-PSTSRLRA------KRPLKCSTCKVIGHNRLTC 857
EG A+ L + RP G P R+RA KR + CS C GH R TC
Sbjct: 93 EGDENAAKADQVLINPPKSLRPPGRPRKKRVRAEDRGRVKRVVHCSRCNQTGHFRTTC 266
>BP049972
Length = 496
Score = 29.3 bits (64), Expect = 2.8
Identities = 19/67 (28%), Positives = 26/67 (38%), Gaps = 6/67 (8%)
Frame = -1
Query: 797 LENKNNPSVPEGVTGTPSANAGLTTHRRRGRPCGPSTSRLRA------KRPLKCSTCKVI 850
+E+ N E + S + + P P R+RA KR + CS C
Sbjct: 394 IEDANANKAVEDANASQSIEIVINPPKSLRPPGRPRKKRVRAEDRGRVKRVVHCSRCNQT 215
Query: 851 GHNRLTC 857
GH R TC
Sbjct: 214 GHFRTTC 194
>TC11650 similar to GB|AAP47462.1|32331869|AY164888 RTNLB1 {Arabidopsis
thaliana;}, partial (7%)
Length = 522
Score = 28.9 bits (63), Expect = 3.6
Identities = 21/82 (25%), Positives = 35/82 (42%), Gaps = 18/82 (21%)
Frame = -2
Query: 409 FDATYKKNKYRRPLVIFSCVNHRNQTTVVGGAVI------------------GNEKEETY 450
FD +Y++N + P F +H N ++ V+ +E EE
Sbjct: 359 FDKSYQRNPFPTPKSQFW--DHSNDNRIIAWWVVKSKMKIIKRDKKVETIIMDDEGEEGG 186
Query: 451 VWLLEQLLEAMDKKSPTTVITD 472
WLL L + KKSP++++ D
Sbjct: 185 AWLLVLPLNIIPKKSPSSLLED 120
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.351 0.156 0.561
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,389,922
Number of Sequences: 28460
Number of extensions: 247350
Number of successful extensions: 2436
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 2402
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2435
length of query: 862
length of database: 4,897,600
effective HSP length: 98
effective length of query: 764
effective length of database: 2,108,520
effective search space: 1610909280
effective search space used: 1610909280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0476b.3


