

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.9
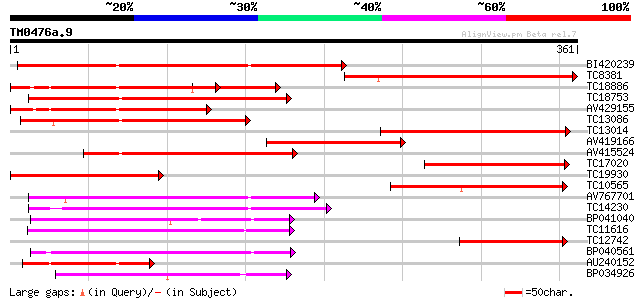
(361 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI420239 286 3e-78
TC8381 207 3e-54
TC18886 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/h... 182 1e-46
TC18753 168 1e-42
AV429155 166 7e-42
TC13086 similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/hydrolas... 152 1e-37
TC13014 150 2e-37
AV419166 150 3e-37
AV415524 148 2e-36
TC17020 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/h... 137 3e-33
TC19930 weakly similar to UP|Q9AX26 (Q9AX26) GDSL-motif lipase/h... 127 4e-30
TC10565 124 2e-29
AV767701 122 1e-28
TC14230 118 2e-27
BP041040 114 2e-26
TC11616 similar to UP|Q9SVU5 (Q9SVU5) Proline-rich APG-like prot... 111 2e-25
TC12742 similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/hydrolas... 109 6e-25
BP040561 109 8e-25
AU240152 108 1e-24
BP034926 104 2e-23
>BI420239
Length = 621
Score = 286 bits (733), Expect = 3e-78
Identities = 139/209 (66%), Positives = 176/209 (83%)
Frame = +1
Query: 6 KTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGI 65
+T LVL+L++++VA ++ + SQVPC+F+FGDSLSDSGNNNNL T AK ++LPYGI
Sbjct: 1 RTLLVLALVILLVAIAIRQYCLQRNSQVPCLFIFGDSLSDSGNNNNLETDAKVDYLPYGI 180
Query: 66 DFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESG 125
DFP T PTGRY+NGRN IDKI ++LG ++FIPPFANL+GSDILKGVNYASGSAGIRKESG
Sbjct: 181 DFP-TGPTGRYTNGRNAIDKITELLGLEDFIPPFANLSGSDILKGVNYASGSAGIRKESG 357
Query: 126 SQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNV 185
+ LG N+N+GLQ+ H AIVS+I+ RL G KA ++L++CLYYVNIGTND+EQNYFLP++
Sbjct: 358 TNLGTNINMGLQIYFHMAIVSQISARL-GFHKAKRHLSKCLYYVNIGTNDYEQNYFLPDL 534
Query: 186 FNTSRMYTPKQYAKALIQQLSHYLQTLHH 214
F+TS YTP++YAK LI +LSHYLQ L +
Sbjct: 535 FDTSSKYTPEEYAKDLINRLSHYLQILRN 621
>TC8381
Length = 844
Score = 207 bits (526), Expect = 3e-54
Identities = 100/151 (66%), Positives = 123/151 (81%), Gaps = 3/151 (1%)
Frame = +3
Query: 214 HFGARKSVLVGLDRLGCIPK--LLVNGSCVEEKNVATFLFNDQLKSLVDRLNKKILMDSK 271
HFGARK+++VG+DRLGCIPK L NGSC+E++NVA FLFNDQLK+LVDR N KIL DSK
Sbjct: 3 HFGARKTIMVGMDRLGCIPKARLTNNGSCIEKENVAAFLFNDQLKALVDRYNHKILPDSK 182
Query: 272 FIFINSTAIIHDKSVGFTVTHHGCCPTN-EKGKCIRDGIPCQNRHEYVFWDGIHTTEAAN 330
FIFINSTAIIHD+S GFT+T CC N +G C+ + PCQNR +Y FWDGIHTTEAAN
Sbjct: 183 FIFINSTAIIHDQSHGFTITDAACCQLNTTRGVCLPNLTPCQNRSQYKFWDGIHTTEAAN 362
Query: 331 LVTAITSYNSSNPAIAYPTNIKHLIQSNTIN 361
++TA SY++S+P IA+P NI+ L+ SN+IN
Sbjct: 363 ILTATVSYSTSDPNIAHPMNIQKLLSSNSIN 455
>TC18886 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif
lipase/hydrolase-like protein, partial (23%)
Length = 547
Score = 182 bits (461), Expect = 1e-46
Identities = 96/139 (69%), Positives = 112/139 (80%), Gaps = 5/139 (3%)
Frame = +1
Query: 1 MGCESKTWLVLSLLVVMVASIMQHSTVLGESQ-VPCIFVFGDSLSDSGNNNNLPTSAKAN 59
M CE K WLVLSL V++ + MQHS VLG SQ VPC+FVFGDSL+DSGNNNNLPT +KAN
Sbjct: 40 MACEIKAWLVLSL--VLMVACMQHS-VLGNSQAVPCLFVFGDSLADSGNNNNLPTLSKAN 210
Query: 60 FLPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYAS---- 115
FLPYGIDFP T PTGRY+NG NPIDK+AQ+LGF++FIPPFANL+GSDILKGVNYAS
Sbjct: 211 FLPYGIDFP-TGPTGRYTNGLNPIDKLAQILGFEKFIPPFANLSGSDILKGVNYASAFSR 387
Query: 116 GSAGIRKESGSQLGHNVNL 134
SAG R + G + H + +
Sbjct: 388 NSAGNRNQFGYKCQHGITI 444
Score = 80.9 bits (198), Expect = 3e-16
Identities = 38/56 (67%), Positives = 45/56 (79%)
Frame = +2
Query: 117 SAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIG 172
SAGIR+E+G+ LG NVN+GLQL HHR IVS+I+ +LGG KA YL QCLYYV IG
Sbjct: 380 SAGIRQETGTNLGTNVNMGLQLQHHRTIVSQISTKLGGFHKAVNYLTQCLYYVYIG 547
>TC18753
Length = 562
Score = 168 bits (425), Expect = 1e-42
Identities = 83/168 (49%), Positives = 118/168 (69%), Gaps = 1/168 (0%)
Frame = +3
Query: 13 LLVVMVASIMQHSTVLG-ESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATF 71
+LV++V +T +G E QVPC F FGDSL D+GNNN L + AKAN+ PYGIDFP
Sbjct: 60 VLVLIVCLWSTTTTGVGAEPQVPCFFSFGDSLVDNGNNNRLSSLAKANYRPYGIDFPGG- 236
Query: 72 PTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKESGSQLGHN 131
PTGR+SNG+ +D I ++LGF +IPP+A G DIL+GVNYAS +AGIR+E+G QLG
Sbjct: 237 PTGRFSNGKTSVDVIGELLGFGSYIPPYATTRGQDILRGVNYASAAAGIREETGQQLGGR 416
Query: 132 VNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQN 179
++ Q+ +++ VS++ + LG + A YL++C+Y + IG+ND+ N
Sbjct: 417 ISFRGQVQNYQRTVSQLINLLGDENTAANYLSKCIYSIGIGSNDYLNN 560
>AV429155
Length = 393
Score = 166 bits (419), Expect = 7e-42
Identities = 83/128 (64%), Positives = 104/128 (80%)
Frame = +3
Query: 1 MGCESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANF 60
M CE+KTWL L LL+ +VA+ MQHS V G+ +VPC+F+FGDSLSD+GNNNNL TS K+N+
Sbjct: 18 MPCETKTWLFLPLLL-LVANCMQHS-VHGKPEVPCLFIFGDSLSDNGNNNNLVTSTKSNY 191
Query: 61 LPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGI 120
PYGIDFP T PTGR++NG +D I+Q+LGF+ FIPPFAN + SDILKGVNYAS +AGI
Sbjct: 192 KPYGIDFP-TGPTGRFTNGPTTVDIISQLLGFENFIPPFANTSDSDILKGVNYASAAAGI 368
Query: 121 RKESGSQL 128
ESG+ +
Sbjct: 369 CVESGTHM 392
>TC13086 similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/hydrolase-like
protein, partial (35%)
Length = 524
Score = 152 bits (383), Expect = 1e-37
Identities = 77/149 (51%), Positives = 108/149 (71%), Gaps = 3/149 (2%)
Frame = +3
Query: 8 WLVLSLL-VVMVASIMQHST--VLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYG 64
W+V++L+ VVMV + ST V + QVPC F+FGDSL D+GNNN L + AKAN+LPYG
Sbjct: 63 WIVMNLVAVVMVVCLGLWSTTRVGADPQVPCYFIFGDSLVDNGNNNQLNS*AKANYLPYG 242
Query: 65 IDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDILKGVNYASGSAGIRKES 124
IDF PTGR+SNG+ +D +A++LGF +IPP++ G DILKGVNYAS +AGIR+E+
Sbjct: 243 IDFGGG-PTGRFSNGKTTVDVVAELLGFDSYIPPYSTARGQDILKGVNYASAAAGIREET 419
Query: 125 GSQLGHNVNLGLQLLHHRAIVSRIAHRLG 153
G QLG ++ Q+ +++ VS++ + LG
Sbjct: 420 GQQLGGRISFSGQVENYQRTVSQVVNLLG 506
>TC13014
Length = 485
Score = 150 bits (380), Expect = 2e-37
Identities = 71/121 (58%), Positives = 86/121 (70%)
Frame = +2
Query: 237 NGSCVEEKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSVGFTVTHHGCC 296
NG+CVEE+N A +FN++LKS V + N K DSKFIFINSTA S+GF V + CC
Sbjct: 20 NGACVEEENAAAIIFNNKLKSSVPKFNPKFSADSKFIFINSTAGSLGNSLGFKVLNASCC 199
Query: 297 PTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIAYPTNIKHLIQ 356
T G CIRD IPCQNR+EY+FWD H T+AAN+ TA SYN+SNPA YP +IK L+Q
Sbjct: 200 KTGAHGLCIRDQIPCQNRNEYIFWDEFHPTKAANIFTAFASYNASNPAFTYPIDIKPLVQ 379
Query: 357 S 357
S
Sbjct: 380 S 382
>AV419166
Length = 270
Score = 150 bits (379), Expect = 3e-37
Identities = 67/89 (75%), Positives = 78/89 (87%)
Frame = +3
Query: 164 QCLYYVNIGTNDFEQNYFLPNVFNTSRMYTPKQYAKALIQQLSHYLQTLHHFGARKSVLV 223
QCLYYV IGTND+EQNYFLP++FNTSR YTP+QYAK L QLSHYL+ LHH GARK+V+V
Sbjct: 3 QCLYYVYIGTNDYEQNYFLPDLFNTSRTYTPEQYAKVLTHQLSHYLKALHHVGARKTVVV 182
Query: 224 GLDRLGCIPKLLVNGSCVEEKNVATFLFN 252
LDRLGCIPK+ VNGSC+E++N A FLFN
Sbjct: 183 SLDRLGCIPKVFVNGSCIEKQNAAAFLFN 269
>AV415524
Length = 411
Score = 148 bits (373), Expect = 2e-36
Identities = 64/136 (47%), Positives = 104/136 (76%)
Frame = +2
Query: 48 NNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFANLNGSDI 107
+NN L + A+A+++PYGIDFP P+GR+SNG+ +D IA++LGF +FIPP+ + +G DI
Sbjct: 2 SNNALRSLARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDI 178
Query: 108 LKGVNYASGSAGIRKESGSQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLY 167
L+GVN+AS +AGIR+E+G QLG ++ Q+ ++++ VS++ + LG D+A YL++C+Y
Sbjct: 179 LRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIY 358
Query: 168 YVNIGTNDFEQNYFLP 183
+ +G+ND+ NYF+P
Sbjct: 359 SIGLGSNDYLNNYFMP 406
>TC17020 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif
lipase/hydrolase-like protein, partial (8%)
Length = 604
Score = 137 bits (345), Expect = 3e-33
Identities = 64/92 (69%), Positives = 75/92 (80%)
Frame = +3
Query: 265 KILMDSKFIFINSTAIIHDKSVGFTVTHHGCCPTNEKGKCIRDGIPCQNRHEYVFWDGIH 324
K +SK++FIN+TAIIHDKS GFTVT CCPTN+ G C D PCQNR+EYVFWDGIH
Sbjct: 3 KPFTNSKYVFINTTAIIHDKSQGFTVTEKVCCPTNKDGVCNPDQTPCQNRNEYVFWDGIH 182
Query: 325 TTEAANLVTAITSYNSSNPAIAYPTNIKHLIQ 356
+TEAANLVTA SY++SN AIA+PTNIK L+Q
Sbjct: 183 STEAANLVTATISYSTSNTAIAHPTNIKKLVQ 278
>TC19930 weakly similar to UP|Q9AX26 (Q9AX26) GDSL-motif
lipase/hydrolase-like protein, partial (11%)
Length = 329
Score = 127 bits (318), Expect = 4e-30
Identities = 59/99 (59%), Positives = 75/99 (75%), Gaps = 1/99 (1%)
Frame = +2
Query: 1 MGCESKTWLVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANF 60
M C KTWL+++L+V++VA + V +QVPC+FVFGDSLSDSGNNNNL T AK +
Sbjct: 32 MACGIKTWLIIALVVLLVAIAIMQIFVQRNTQVPCLFVFGDSLSDSGNNNNLETLAKVAY 211
Query: 61 LPYGIDFP-ATFPTGRYSNGRNPIDKIAQMLGFQEFIPP 98
PYGIDFP PTGRYSNGR +DK+ ++LGF++FIPP
Sbjct: 212 PPYGIDFPTGPTPTGRYSNGRTAVDKLTELLGFEDFIPP 328
>TC10565
Length = 616
Score = 124 bits (312), Expect = 2e-29
Identities = 63/116 (54%), Positives = 80/116 (68%), Gaps = 3/116 (2%)
Frame = +2
Query: 243 EKNVATFLFNDQLKSLVDRLNKKILMDSKFIFINSTAIIHDKSV--GFTVTHHGCCPTNE 300
E+N A F+FND+LKSLV+ N K+ SKFIFIN+T+I + ++ G +VT CC
Sbjct: 2 EENAAAFIFNDKLKSLVELFNNKLSATSKFIFINTTSIDQENTLVSGISVTDAACCSPGL 181
Query: 301 KGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYN-SSNPAIAYPTNIKHLI 355
G+CI D IPC NR +YVFWD HT EA NL+TAI SY+ SSN A YP +IKHL+
Sbjct: 182 LGECIPDEIPCYNRSDYVFWDEFHTPEAWNLLTAIKSYDTSSNQAFTYPLDIKHLV 349
>AV767701
Length = 622
Score = 122 bits (305), Expect = 1e-28
Identities = 72/192 (37%), Positives = 107/192 (55%), Gaps = 7/192 (3%)
Frame = +3
Query: 13 LLVVMVASIMQHSTVLGESQVP------CIFVFGDSLSDSGNNNNLPTSAKANFLPYGID 66
+L+V+V I + +S P FVFGDSL DSGNNN L T+A+A+ PYGID
Sbjct: 48 ILLVVVGVIFPGAEAKSKSSKPKPKPSRAFFVFGDSLVDSGNNNYLATTARADSPPYGID 227
Query: 67 FPATFPTGRYSNGRNPIDKIAQMLGFQEFIPPFA-NLNGSDILKGVNYASGSAGIRKESG 125
+P PTGR+SNG N D I+Q LG + +P + L G+ +L G N+AS GI ++G
Sbjct: 228 YPTRRPTGRFSNGLNIPDLISQQLGAESVLPYLSPQLRGNKLLLGANFASAGIGILNDTG 407
Query: 126 SQLGHNVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNV 185
+Q + + + QL + R+A ++ G+ K +++ L + +G NDF NY+L
Sbjct: 408 TQFLNIIRMYRQLDYFEEYQHRLASQI-GVTKTKALVDKALVLITVGGNDFVNNYYLVPY 584
Query: 186 FNTSRMYTPKQY 197
SR Y+ Y
Sbjct: 585 SARSRQYSLPDY 620
>TC14230
Length = 740
Score = 118 bits (295), Expect = 2e-27
Identities = 71/194 (36%), Positives = 107/194 (54%), Gaps = 1/194 (0%)
Frame = +1
Query: 13 LLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFP 72
+LV+ ++S+ T FVFGDSL DSGNN+ L T+A+A+ PYGIDFP P
Sbjct: 91 VLVLFLSSVSAQHT-------RAFFVFGDSLVDSGNNDFLATTARADAPPYGIDFPTHRP 249
Query: 73 TGRYSNGRNPIDKIAQMLGFQEFIPPFAN-LNGSDILKGVNYASGSAGIRKESGSQLGHN 131
TGR+SNG N D I+ LG + +P + L G +L G N+AS GI ++G Q H
Sbjct: 250 TGRFSNGLNIPDLISLELGLEPTLPYLSPLLVGEKLLIGANFASAGIGILNDTGIQFLHI 429
Query: 132 VNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFLPNVFNTSRM 191
+++ QL + R++ + G + +N+ L+ + +G NDF NY+L SR
Sbjct: 430 IHIEKQLKLFQQYQKRVSAHI-GREGTRNLVNRALFLITLGGNDFVNNYYLVPYSARSRQ 606
Query: 192 YTPKQYAKALIQQL 205
Y+ Y + +I +L
Sbjct: 607 YSLPDYVRYIISEL 648
>BP041040
Length = 550
Score = 114 bits (286), Expect = 2e-26
Identities = 67/170 (39%), Positives = 97/170 (56%), Gaps = 2/170 (1%)
Frame = +3
Query: 14 LVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPT 73
L + + I H + +VP I VFGDS DSGNNN +PT AK+NF PYG DFP PT
Sbjct: 21 LWLFLIEIFLHISTSSSGKVPAIIVFGDSSVDSGNNNFIPTMAKSNFEPYGRDFPDGNPT 200
Query: 74 GRYSNGRNPIDKIAQMLGFQEFIPPFAN--LNGSDILKGVNYASGSAGIRKESGSQLGHN 131
GR+SNGR D I++ G + IP + + + SD GV +AS G S S +
Sbjct: 201 GRFSNGRIAPDFISEAFGLKPTIPAYLDPAYSISDFASGVCFASAGTGY-DNSTSNVADV 377
Query: 132 VNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYF 181
+ L ++ +++ ++ LG +KA + + + LY V+IGTNDF +NY+
Sbjct: 378 IPLWKEVEYYKDYRQKLVAYLGD-EKANEIVKEALYLVSIGTNDFLENYY 524
>TC11616 similar to UP|Q9SVU5 (Q9SVU5) Proline-rich APG-like protein,
partial (41%)
Length = 569
Score = 111 bits (278), Expect = 2e-25
Identities = 66/171 (38%), Positives = 96/171 (55%), Gaps = 1/171 (0%)
Frame = +2
Query: 12 SLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATF 71
SL +V + S G FVFGDSL D+GNNN L T+A+A+ PYGID P
Sbjct: 59 SLACYIVLVVALASGFRGAEAARAFFVFGDSLVDNGNNNYLATTARADSPPYGIDTPNGR 238
Query: 72 PTGRYSNGRNPIDKIAQMLGFQEFIPPFA-NLNGSDILKGVNYASGSAGIRKESGSQLGH 130
PTGR+SNGRN D I++ LG + +P + LNG ++L G N+AS GI ++G Q +
Sbjct: 239 PTGRFSNGRNIPDFISEQLGAEPTLPYLSPELNGDNLLVGANFASAGIGILNDTGIQFIN 418
Query: 131 NVNLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYF 181
+ + QL + R++ L G ++ + +N L + +G NDF NY+
Sbjct: 419 IIRIFRQLEYFDEYQQRVS-ALIGPEETQRLVNGALVLITLGGNDFVNNYY 568
>TC12742 similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/hydrolase-like
protein, partial (6%)
Length = 590
Score = 109 bits (273), Expect = 6e-25
Identities = 48/69 (69%), Positives = 58/69 (83%)
Frame = +3
Query: 287 GFTVTHHGCCPTNEKGKCIRDGIPCQNRHEYVFWDGIHTTEAANLVTAITSYNSSNPAIA 346
GF T+ CC TNE+GKCI +G PCQNR EYVFWDGIHT+EAAN+VTA SY++S+ AIA
Sbjct: 6 GFKFTNAPCCTTNERGKCILNGTPCQNRTEYVFWDGIHTSEAANIVTATISYSTSDSAIA 185
Query: 347 YPTNIKHLI 355
+PTNIKHL+
Sbjct: 186 HPTNIKHLV 212
>BP040561
Length = 515
Score = 109 bits (272), Expect = 8e-25
Identities = 64/170 (37%), Positives = 99/170 (57%), Gaps = 1/170 (0%)
Frame = +1
Query: 14 LVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPT 73
L V+V S+ + +Q FVFGDSL D+GNN+ L T+A+A+ PYGIDFP PT
Sbjct: 16 LCVIVTSLFM---AVDSTQTRAFFVFGDSLVDNGNNDFLATTARADAPPYGIDFPTHKPT 186
Query: 74 GRYSNGRNPIDKIAQMLGFQEFIPPFAN-LNGSDILKGVNYASGSAGIRKESGSQLGHNV 132
GR+SNG N D I++ LG + +P + L G +L G N+AS GI ++G Q + +
Sbjct: 187 GRFSNGLNIPDLISEQLGLEPTMPYLSPLLMGEKLLVGANFASAGIGILNDTGFQFLNII 366
Query: 133 NLGLQLLHHRAIVSRIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQNYFL 182
++ QL + R++ ++G + + +N+ L + +G NDF NY+L
Sbjct: 367 HMYKQLKLFQHYQQRLSAQIGP-EGTKKLVNKALVLITLGGNDFVNNYYL 513
>AU240152
Length = 300
Score = 108 bits (271), Expect = 1e-24
Identities = 54/84 (64%), Positives = 67/84 (79%)
Frame = +1
Query: 9 LVLSLLVVMVASIMQHSTVLGESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFP 68
+V+ L+++VA MQHS V GE +VPC+F+FGDSLSD GNNNNLPTS K+N+ PYGIDFP
Sbjct: 55 MVVLPLLLLVAKSMQHS-VHGEPEVPCLFIFGDSLSDDGNNNNLPTSTKSNYNPYGIDFP 231
Query: 69 ATFPTGRYSNGRNPIDKIAQMLGF 92
T PTGR++NG ID I Q+LGF
Sbjct: 232 -TGPTGRFTNGPTAIDIIGQLLGF 300
>BP034926
Length = 607
Score = 104 bits (260), Expect = 2e-23
Identities = 64/153 (41%), Positives = 89/153 (57%), Gaps = 3/153 (1%)
Frame = +2
Query: 30 ESQVPCIFVFGDSLSDSGNNNNLPTSAKANFLPYGIDFPATFPTGRYSNGRNPIDKIAQM 89
+S V CI VFGDS D GNNN L TS K+NF PYG DF + PTGR+ NGR D IA+
Sbjct: 158 KSNVSCILVFGDSSVDPGNNNVLRTSMKSNFPPYGKDFFNSLPTGRFCNGRLATDFIAEA 337
Query: 90 LGFQEFIPPF--ANLNGSDILKGVNYASGSAGIRKESGSQLG-HNVNLGLQLLHHRAIVS 146
LG+++ +P F NL D+ GV++AS + G + + + V+ +Q H I
Sbjct: 338 LGYRQMLPAFLDPNLKVEDLPYGVSFASAATGFDDYTANVVNVLPVSKQIQYFMHYKIHL 517
Query: 147 RIAHRLGGLDKATQYLNQCLYYVNIGTNDFEQN 179
R +L G ++A + L+ V++GTNDF QN
Sbjct: 518 R---KLLGEERAEFIIRNALFIVSMGTNDFLQN 607
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,644,426
Number of Sequences: 28460
Number of extensions: 92053
Number of successful extensions: 573
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 534
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 534
length of query: 361
length of database: 4,897,600
effective HSP length: 91
effective length of query: 270
effective length of database: 2,307,740
effective search space: 623089800
effective search space used: 623089800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0476a.9


