

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0476a.5
(268 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
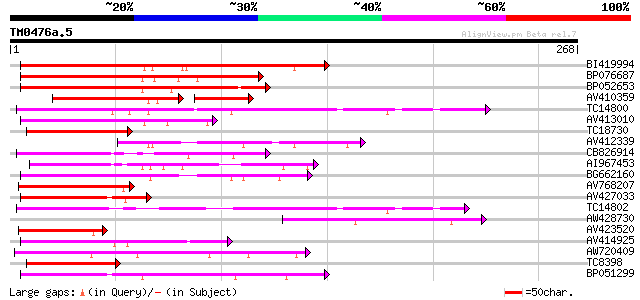
Score E
Sequences producing significant alignments: (bits) Value
BI419994 151 1e-37
BP076687 122 6e-29
BP052653 117 2e-27
AV410359 74 7e-21
TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromo... 84 3e-17
AV413010 82 1e-16
TC18730 similar to UP|Q963J2 (Q963J2) Voltage-dependent calcium ... 80 4e-16
AV412339 74 3e-14
CB826914 73 4e-14
AI967453 71 2e-13
BG662160 68 2e-12
AV768207 66 5e-12
AV427033 66 5e-12
TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (... 65 9e-12
AW428730 64 3e-11
AV423520 63 5e-11
AV414925 62 8e-11
AW720409 62 1e-10
TC8398 weakly similar to UP|Q84KK1 (Q84KK1) S haplotype-specific... 58 2e-09
BP051299 53 6e-08
>BI419994
Length = 549
Score = 151 bits (381), Expect = 1e-37
Identities = 87/164 (53%), Positives = 101/164 (61%), Gaps = 18/164 (10%)
Frame = +2
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCL--- 62
LP ELI +ILLRLPVRSLLRFK VCKSW YLISDPQF KSHF+L A+PTHR+ L
Sbjct: 47 LPDELILQILLRLPVRSLLRFKTVCKSWRYLISDPQFTKSHFNLAASPTHRIFLNNTNGF 226
Query: 63 ----FDLD-SLGDNSAVVTLNLP--PP-----CMSCIQNSLYFLGSCRGFMLLANEYQRV 110
D D SL D S++V L P PP L LGSCRGF+LLAN V
Sbjct: 227 *IESLDTDSSLHDESSLVHLKFPLAPPHPDNHRFGSSVTGLKVLGSCRGFVLLANHQGNV 406
Query: 111 IVWNPSTGFHKQILTSCDFIIGS---LYGFGYDNSTDDYFLVLI 151
+VWNPSTG +++ C ++ LYG GYD S DDY +V I
Sbjct: 407 VVWNPSTGVRRRVPEWCSYLASQCEFLYGIGYDESNDDYLVVFI 538
>BP076687
Length = 389
Score = 122 bits (306), Expect = 6e-29
Identities = 69/127 (54%), Positives = 85/127 (66%), Gaps = 12/127 (9%)
Frame = -3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRC---- 61
LP ELI +ILLRLPVRSLLRFK VCKSW LISDP+F KSH+DL A+PT+R L
Sbjct: 381 LPEELIPQILLRLPVRSLLRFKSVCKSWLSLISDPKFGKSHYDLAASPTYRCLTIANDSE 202
Query: 62 --LFDLDS--LGDNSAVVTLN--LPPPCMSCI--QNSLYFLGSCRGFMLLANEYQRVIVW 113
D+D+ L S+VV L LPPPC + N+++FLG CRGF+++ E V +W
Sbjct: 201 IGSIDVDAPLLNYESSVVKLKYPLPPPCKLSLFSGNTVWFLGYCRGFVIVVYEQGDVFLW 22
Query: 114 NPSTGFH 120
NPSTG H
Sbjct: 21 NPSTGVH 1
>BP052653
Length = 552
Score = 117 bits (293), Expect = 2e-27
Identities = 67/123 (54%), Positives = 82/123 (66%), Gaps = 5/123 (4%)
Frame = +1
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRC-LFD 64
LPH+LI +ILLRLPVRSLLRFK VCKSW LISDPQF KSHFDL AAPTHR L++ F
Sbjct: 187 LPHDLIVQILLRLPVRSLLRFKSVCKSWLSLISDPQFGKSHFDLAAAPTHRCLVKINNFK 366
Query: 65 LDSLGDNSAVV----TLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFH 120
L+SL +++V+ +NL P + + LGSCRGF+ L IVWNPS
Sbjct: 367 LESLDVDASVLDQSSEVNLKFPYTGYVIFHIQILGSCRGFIALVRGGD-AIVWNPSXCVQ 543
Query: 121 KQI 123
++I
Sbjct: 544 RKI 552
>AV410359
Length = 342
Score = 73.6 bits (179), Expect(2) = 7e-21
Identities = 41/72 (56%), Positives = 50/72 (68%), Gaps = 10/72 (13%)
Frame = +2
Query: 21 RSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFD---LDSL-------GD 70
RSL+RFK VCKSW LISDP+F KSHFDL AAPTHR L+R + D ++SL +
Sbjct: 5 RSLIRFKSVCKSWRSLISDPKFGKSHFDLAAAPTHRYLVRKINDSSEIESLDFNASLHDE 184
Query: 71 NSAVVTLNLPPP 82
+SA+V L LP P
Sbjct: 185SSAIVKLTLPFP 220
Score = 42.7 bits (99), Expect(2) = 7e-21
Identities = 17/28 (60%), Positives = 22/28 (77%)
Frame = +1
Query: 88 QNSLYFLGSCRGFMLLANEYQRVIVWNP 115
QN +Y LGSCRGF+++A E V+VWNP
Sbjct: 259 QNPIYILGSCRGFVVVAYERGSVVVWNP 342
>TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromosome 5, TAC
clone:K14B20, partial (7%)
Length = 782
Score = 84.0 bits (206), Expect = 3e-17
Identities = 76/246 (30%), Positives = 116/246 (46%), Gaps = 22/246 (8%)
Frame = +3
Query: 4 LTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHF-----DLNAAPTH-RL 57
L LP ELI EIL LPV+SLL+F+ V K+W ISDPQF K H NA H L
Sbjct: 42 LCLPDELIIEILSWLPVKSLLQFRVVSKTWKSFISDPQFVKLHLLHRLSFRNADFEHTSL 221
Query: 58 LLRCLFD------LDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLL-------- 103
L++C D + S +S + + + SCI + F+G+C G + L
Sbjct: 222 LIKCHTDDFGRPYISSRTVSSLLESPSAIVASRSCI-SGYDFIGTCNGLVSLRKLNYDES 398
Query: 104 -ANEYQRVIVWNPSTGFHKQILTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHA 162
N + +V WNP+T Q GFGYD S+D Y +V ++ ++
Sbjct: 399 NTNNFSQVRFWNPATRTMSQDSPPSWSPRNLHLGFGYDCSSDTYKVVGMIPGL--TMVNV 572
Query: 163 YSVKTNSWDYNHVHV-LYMHLGCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSETSLS 221
Y++ N W + MHL V+++N+L+WL T+ ++++DL + +
Sbjct: 573 YNMGDNCWRTIQISPHAPMHL---QGSAVYVSNTLNWLA---GTNPYFIVSFDLEKEECA 734
Query: 222 EIPLLP 227
+ LP
Sbjct: 735 QQLSLP 752
>AV413010
Length = 416
Score = 82.0 bits (201), Expect = 1e-16
Identities = 52/107 (48%), Positives = 62/107 (57%), Gaps = 14/107 (13%)
Frame = +3
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCL--F 63
LP ELI +ILL+LPV SLLRFK V KSWF IS+PQFAKSHF+L A+PTH L L+ F
Sbjct: 96 LPDELIIQILLQLPVTSLLRFKSVSKSWFSQISNPQFAKSHFNLAASPTHYLFLKSTDEF 275
Query: 64 DLDSLGDNSA----VVTLNLPPPCMSCIQNSLY--------FLGSCR 98
+L S+ LN P NS++ LGSCR
Sbjct: 276 QFQTLDIESSPPFTTAVLNYPHEQPRSGSNSIFCPSITDLEVLGSCR 416
>TC18730 similar to UP|Q963J2 (Q963J2) Voltage-dependent calcium channel
alpha13 subunit (Fragment), partial (5%)
Length = 515
Score = 80.1 bits (196), Expect = 4e-16
Identities = 37/50 (74%), Positives = 42/50 (84%)
Frame = +2
Query: 9 ELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLL 58
ELI ILLRLPVRSLLRFK VCKSW LISDP+F SH+D++A+PTHR L
Sbjct: 230 ELILHILLRLPVRSLLRFKSVCKSWLSLISDPKFGXSHYDISASPTHRCL 379
>AV412339
Length = 420
Score = 73.9 bits (180), Expect = 3e-14
Identities = 60/139 (43%), Positives = 71/139 (50%), Gaps = 22/139 (15%)
Frame = +3
Query: 52 APTHRLLLRCLFD------LD---SLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFML 102
+PTHRLLL + D LD SL D SAVV L PP S LGSCRG L
Sbjct: 3 SPTHRLLLNRINDGSEMESLDVEASLDDPSAVVNLKFPP-------YSGRTLGSCRGLFL 161
Query: 103 LANEYQR-------VIVWNPSTGFHKQILTSCDFIIGS--LYGFGYDNSTDDYFLVLIVL 153
L E +IVWNPSTG +++ D+ GS LYGFGYD S DDY LV+I L
Sbjct: 162 LTKEMSPSGSSPGDLIVWNPSTGAYREFP---DWDSGSHTLYGFGYDPSNDDYLLVVIRL 332
Query: 154 ASMNA----KIHAYSVKTN 168
+ +I + KTN
Sbjct: 333 CEFDPGRTNQITVFPGKTN 389
>CB826914
Length = 575
Score = 73.2 bits (178), Expect = 4e-14
Identities = 53/148 (35%), Positives = 71/148 (47%), Gaps = 28/148 (18%)
Frame = +1
Query: 4 LTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLF 63
+ + +LI EILL LPV+S++RFK VCK W LISDP FA HF AAP+ LF
Sbjct: 160 IDMDRDLIIEILLMLPVKSIVRFKAVCKLWRSLISDPLFASLHFQ-RAAPS------LLF 318
Query: 64 DLDSLGDNSAVVTLNLPPPC-------------------MSCIQNSLYFLGSCRGFMLLA 104
D+ A+ T++L P +S +N + GSCRGF+L+
Sbjct: 319 -----ADSHAIRTIDLEGPLQSHRVSQPINCHFLSNYHDLSLPRNCISIHGSCRGFLLIT 483
Query: 105 ---------NEYQRVIVWNPSTGFHKQI 123
+ +WNPST HK I
Sbjct: 484 WISNIGYGHPWNDSLYLWNPSTHVHKPI 567
>AI967453
Length = 456
Score = 70.9 bits (172), Expect = 2e-13
Identities = 60/152 (39%), Positives = 81/152 (52%), Gaps = 15/152 (9%)
Frame = +1
Query: 10 LIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRC-----LFD 64
LI EILLRLPV+SL+RFK VCK W LISDP FA SHF+ AAP RLL+R D
Sbjct: 16 LITEILLRLPVKSLVRFKAVCKFWRSLISDPHFATSHFE-RAAP--RLLIRTDHGIRTMD 186
Query: 65 L-DSLGDN--SAVVTLNLP---PPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTG 118
L +SL + S ++ + P P + ++ S +L S R + +WNPST
Sbjct: 187 LEESLHPDRISELIDYDFPYTVPYIVVRVELSANWL*SSRS----------LSIWNPSTH 336
Query: 119 FHKQILTSCD-FIIGSLYGFGY---DNSTDDY 146
HK + + + +Y + Y +S DDY
Sbjct: 337 VHKPLPVDANAYEYDHMYPYEYGLVTSSKDDY 432
>BG662160
Length = 445
Score = 67.8 bits (164), Expect = 2e-12
Identities = 51/149 (34%), Positives = 73/149 (48%), Gaps = 11/149 (7%)
Frame = +2
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLFDL 65
LP EL EIL LPV+SL+RF+CV KSW +ISD QF K H +++ T L L
Sbjct: 23 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQSL 202
Query: 66 -DSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLL-ANEYQR-------VIVWNPS 116
S + + T+ +P + F G+C G + L +++Y R V +WNP+
Sbjct: 203 ITSPRKSRSASTIAIP--------DDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPA 358
Query: 117 TGFHKQILTS--CDFIIGSLYGFGYDNST 143
Q + S +GF YD+ST
Sbjct: 359 MRSMSQRSPPLYLPIVYDSYFGFDYDSST 445
>AV768207
Length = 374
Score = 66.2 bits (160), Expect = 5e-12
Identities = 34/57 (59%), Positives = 41/57 (71%), Gaps = 2/57 (3%)
Frame = +1
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAA--PTHRLLL 59
TLP EL EIL RLPV+SLL+ +CVCKSW LISDP+FAK+H ++ HRL L
Sbjct: 70 TLPFELGVEILCRLPVKSLLQLRCVCKSWKSLISDPKFAKNHLRCSSTDFTRHRLFL 240
>AV427033
Length = 420
Score = 66.2 bits (160), Expect = 5e-12
Identities = 36/66 (54%), Positives = 46/66 (69%), Gaps = 4/66 (6%)
Frame = +1
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAP----THRLLLRC 61
LP +L+ EIL RLPV SLL+F+CVCKSW LISDP+FAK+H L+ +P H L+
Sbjct: 124 LPFDLVVEILCRLPVNSLLQFRCVCKSWNSLISDPKFAKNH--LHCSPPDFTRHHLMGSD 297
Query: 62 LFDLDS 67
+ D DS
Sbjct: 298 IEDEDS 315
>TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (SLF)-S4
protein, partial (10%)
Length = 551
Score = 65.5 bits (158), Expect = 9e-12
Identities = 64/215 (29%), Positives = 93/215 (42%), Gaps = 1/215 (0%)
Frame = +3
Query: 4 LTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRCLF 63
L LP ELI EIL LPV+SLL+F+ V K+W ISDPQF K H HRL R
Sbjct: 15 LCLPDELIIEILSWLPVKSLLQFRVVSKTWKSFISDPQFVKLHL------LHRLSFR--- 167
Query: 64 DLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLANEYQRVIVWNPSTGFHKQI 123
N+ +L C + Y + V WNP+T Q
Sbjct: 168 -------NADFEHTSLLIKCHTDDFGRPYI------------SSRTVRFWNPATRTMSQD 290
Query: 124 LTSCDFIIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHAYSVKTNSWDYNHVHV-LYMHL 182
GFGYD S+D Y +V ++ ++ Y++ N W + MHL
Sbjct: 291 SPPSWSPRNLHLGFGYDCSSDTYKVVGMIPGL--TMVNVYNMGDNCWRTIQISPHAPMHL 464
Query: 183 GCDYRHGVFLNNSLHWLVISKVTSLQVVIAYDLSE 217
V+++N+L+WL T+ ++++DL +
Sbjct: 465 ---QGSAVYVSNTLNWLA---GTNPYFIVSFDLEK 551
>AW428730
Length = 405
Score = 63.9 bits (154), Expect = 3e-11
Identities = 35/103 (33%), Positives = 52/103 (49%), Gaps = 7/103 (6%)
Frame = +3
Query: 130 IIGSLYGFGYDNSTDDYFLVLIVLASMNAKIHA-----YSVKTNSWDYNHVHVLYMHLGC 184
+ L+GFGYD+STDDY +V + + H +S++ N W Y L
Sbjct: 27 VFSHLFGFGYDSSTDDYLVVRVPVTDCYQPTHLPDVQFFSLRANMWKYTEGVDLPPLTTI 206
Query: 185 DYRHGVFLNNSLHWLVISKVTSL--QVVIAYDLSETSLSEIPL 225
D HG+ N ++HW V + V + +IA+DL E L EIP+
Sbjct: 207 DLCHGLLFNEAIHWAVSNWVDGVTNMFIIAFDLMEKRLLEIPM 335
>AV423520
Length = 468
Score = 63.2 bits (152), Expect = 5e-11
Identities = 30/43 (69%), Positives = 36/43 (82%), Gaps = 1/43 (2%)
Frame = +3
Query: 5 TLPHELIFEILLRLPVRSLLRFKCVCKSWFYLIS-DPQFAKSH 46
TLP EL+ EIL RLPV+SLL+F+CVCKSW LIS DP+FA+ H
Sbjct: 162 TLPFELVVEILCRLPVKSLLQFRCVCKSWNSLISGDPKFARKH 290
>AV414925
Length = 414
Score = 62.4 bits (150), Expect = 8e-11
Identities = 42/104 (40%), Positives = 53/104 (50%), Gaps = 4/104 (3%)
Frame = +1
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFD---LNAAPT-HRLLLRC 61
+P E+I +IL RLPV SLLRF+ + KSW LI F K H + N P L+LR
Sbjct: 97 IPVEVIADILSRLPVTSLLRFRSISKSWRSLIDSKHFMKLHLNNSLTNPNPNLTNLILRH 276
Query: 62 LFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLAN 105
DL S +NL P M C N + LGSC G + + N
Sbjct: 277 NTDLYRADFPSIGAAVNLNHPLM-CYSNRINILGSCHGLLCICN 405
>AW720409
Length = 575
Score = 62.0 bits (149), Expect = 1e-10
Identities = 52/163 (31%), Positives = 73/163 (43%), Gaps = 23/163 (14%)
Frame = +3
Query: 3 KLTLPHELIFEILLRLPVRSLLRFKCVCKSWFYLI-SDPQFAKSHFDLNAAPTHRLLL-- 59
+ LP EL EIL LPV++L+RF CV KSW LI D F K H D + ++++L
Sbjct: 84 EFVLPFELWIEILSWLPVKTLMRFSCVSKSWKSLIYQDRDFKKLHLDRSPKNNNQVILTL 263
Query: 60 -RCLFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYF--------------LGSCRGFMLLA 104
+ LF + L P S I + + F +GSC G + L
Sbjct: 264 QKPLFSGRTFFPFPVRRLLQDQEPSSSSIIDDIPFEEEDEDEEDNYHDTIGSCNGLVCLY 443
Query: 105 NEYQRVI---VWNPSTGFHKQILTSCDFIIGSL--YGFGYDNS 142
+ +WNP+T F D +IGS + FGYD+S
Sbjct: 444 GINDHGLWFRLWNPATRFRFHKSPPLDAVIGSTLHFAFGYDHS 572
>TC8398 weakly similar to UP|Q84KK1 (Q84KK1) S haplotype-specific F-box
protein a, partial (9%)
Length = 1602
Score = 57.8 bits (138), Expect = 2e-09
Identities = 27/44 (61%), Positives = 34/44 (76%)
Frame = +3
Query: 9 ELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAA 52
ELI+EIL LPV+SL+RF+CVCKSW IS+PQF K H ++A
Sbjct: 87 ELIWEILSLLPVKSLVRFRCVCKSWKLTISNPQFMKLHLRRSSA 218
>BP051299
Length = 539
Score = 52.8 bits (125), Expect = 6e-08
Identities = 43/156 (27%), Positives = 71/156 (44%), Gaps = 10/156 (6%)
Frame = +2
Query: 6 LPHELIFEILLRLPVRSLLRFKCVCKSWFYLISDPQFAKSHFDLNAAPTHRLLLRC---- 61
LP E+ EIL RLP ++L++ VCKSW LI+ F H N +P+ LL C
Sbjct: 5 LPQEIWAEILHRLPPKTLVKCTSVCKSWRSLITSTSFISLH--RNHSPSSLLLQLCDERA 178
Query: 62 --LFDLDSLGDNSAVVTLNLPPPCMSCIQNSLYFLGSCRGFMLLAN--EYQRVIVWNPST 117
F SL + ++ + S +G C G + + + + +I+ NPS
Sbjct: 179 IPNFIHYSLRRDDPFLSESSSLRLPSSFNREFSVVGICHGLVCITSTENCRDLIICNPSL 358
Query: 118 GFHKQILTSCDF--IIGSLYGFGYDNSTDDYFLVLI 151
H + D+ + + FG+D+ +DY ++ I
Sbjct: 359 RLHITLPEPSDYPSLHSASVAFGFDSRNNDYKVIRI 466
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.327 0.142 0.456
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,185,243
Number of Sequences: 28460
Number of extensions: 106195
Number of successful extensions: 791
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 765
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 768
length of query: 268
length of database: 4,897,600
effective HSP length: 89
effective length of query: 179
effective length of database: 2,364,660
effective search space: 423274140
effective search space used: 423274140
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0476a.5


