

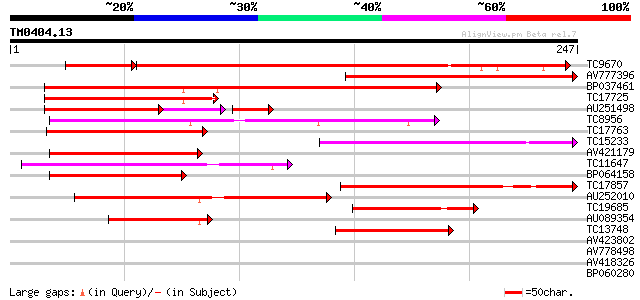
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0404.13
(247 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%) 281 1e-89
AV777396 204 1e-53
BP037461 156 3e-39
TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, part... 110 2e-25
AU251498 102 3e-25
TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, pa... 109 4e-25
TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor,... 93 4e-20
TC15233 similar to UP|AAS45681 (AAS45681) AGAMOUS-like protein (... 88 2e-18
AV421179 84 3e-17
TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, pa... 83 5e-17
BP064158 81 2e-16
TC17857 similar to UP|Q8VWZ3 (Q8VWZ3) C-type MADS box protein, p... 79 7e-16
AU252010 75 8e-15
TC19685 similar to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS-2,... 53 6e-08
AU089354 45 2e-05
TC13748 similar to UP|Q39401 (Q39401) MADS5 protein, partial (28%) 45 2e-05
AV423802 27 3.3
AV778498 27 3.3
AV418326 27 4.3
BP060280 25 9.5
>TC9670 homologue to UP|O64958 (O64958) CUM1, partial (65%)
Length = 1060
Score = 281 bits (720), Expect(2) = 1e-89
Identities = 151/195 (77%), Positives = 171/195 (87%), Gaps = 6/195 (3%)
Frame = +3
Query: 56 CDAEVALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLR 115
CDAEVALIVFS+RGRLYEYANNSVKA+I+RYKKACSDSSG GS S ANAQFYQQEA KLR
Sbjct: 93 CDAEVALIVFSSRGRLYEYANNSVKATIDRYKKACSDSSGAGSASEANAQFYQQEADKLR 272
Query: 116 VQISNLQNHNRQMLGEALSNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREI 175
VQISNLQN+NRQM+GE+L +MNA++LKNLETKLEKGISRIRSKKNE+LFAEIEYMQKREI
Sbjct: 273 VQISNLQNNNRQMMGESLGSMNAKELKNLETKLEKGISRIRSKKNELLFAEIEYMQKREI 452
Query: 176 DLHNSNQLLRAKIAESDERKNHNFNMLPG-TTNFESL----QQSQQPFDSRGFFQVTGLQ 230
DLHN+NQLLRAKIAES ER + N ++L G T+N+ES+ QQ QQ FDSRG+FQVTGLQ
Sbjct: 453 DLHNNNQLLRAKIAES-ERNHPNLSILAGSTSNYESMQSQQQQQQQQFDSRGYFQVTGLQ 629
Query: 231 P-NNNQCARQDQISL 244
P + +R D ISL
Sbjct: 630 PTTHTPYSRPDPISL 674
Score = 64.7 bits (156), Expect(2) = 1e-89
Identities = 31/31 (100%), Positives = 31/31 (100%)
Frame = +1
Query: 25 IKRIENTTNRQVTFCKRRNGLLKKAYELSVL 55
IKRIENTTNRQVTFCKRRNGLLKKAYELSVL
Sbjct: 1 IKRIENTTNRQVTFCKRRNGLLKKAYELSVL 93
>AV777396
Length = 577
Score = 204 bits (519), Expect = 1e-53
Identities = 101/101 (100%), Positives = 101/101 (100%)
Frame = -2
Query: 147 KLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHNFNMLPGTT 206
KLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHNFNMLPGTT
Sbjct: 576 KLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHNFNMLPGTT 397
Query: 207 NFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
NFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV
Sbjct: 396 NFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 274
>BP037461
Length = 551
Score = 156 bits (395), Expect = 3e-39
Identities = 82/175 (46%), Positives = 122/175 (68%), Gaps = 2/175 (1%)
Frame = +2
Query: 16 RKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY- 74
R+MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALI+FSNRG+LYE+
Sbjct: 2 REMGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFC 181
Query: 75 ANNSVKASIERYKKA-CSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEAL 133
+++S+ ++ERY+K ST A QQE KL+ + LQ R ++GE L
Sbjct: 182 SSSSMLKTLERYQKCNYGAPEANVSTREALELSSQQEYLKLKARYEALQRSQRNLMGEDL 361
Query: 134 SNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKI 188
+N+++L++LE +L+ + +IRS + + + ++ +Q++E L +N+ LR ++
Sbjct: 362 GPLNSKELESLERQLDSSLKQIRSTRTQFMLDQLSDLQRKEHMLSEANRSLRQRL 526
>TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, partial (45%)
Length = 970
Score = 110 bits (275), Expect = 2e-25
Identities = 56/77 (72%), Positives = 68/77 (87%), Gaps = 1/77 (1%)
Frame = +3
Query: 16 RKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEY- 74
R MGRG++E+KRIEN NRQVTF KRRNGLLKKAYELSVLCDAEVALIVFS RG+LYE+
Sbjct: 270 RDMGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIVFSTRGKLYEFC 449
Query: 75 ANNSVKASIERYKKACS 91
+++S+ ++ERY+K CS
Sbjct: 450 SSSSMVKTLERYQK-CS 497
>AU251498
Length = 342
Score = 102 bits (253), Expect(2) = 3e-25
Identities = 50/52 (96%), Positives = 52/52 (99%)
Frame = +3
Query: 16 RKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSN 67
+KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFS+
Sbjct: 39 QKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSS 194
Score = 50.4 bits (119), Expect = 3e-07
Identities = 25/79 (31%), Positives = 42/79 (52%)
Frame = +1
Query: 16 RKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYA 75
R+ G G++ + + + + G +K ++ + RGRLYEY+
Sbjct: 40 RRWGGGRLRSRGLRTQLIAR*HSAREETGF*RKLMSYQCCVMLKLPSLSSPARGRLYEYS 219
Query: 76 NNSVKASIERYKKACSDSS 94
NN+++++IERYKKACSD S
Sbjct: 220 NNNIRSTIERYKKACSDHS 276
Score = 28.9 bits (63), Expect(2) = 3e-25
Identities = 12/18 (66%), Positives = 16/18 (88%)
Frame = +2
Query: 98 STSGANAQFYQQEAAKLR 115
+T+ NAQ+YQQE+AKLR
Sbjct: 287 TTTEINAQYYQQESAKLR 340
>TC8956 similar to UP|Q9ZS28 (Q9ZS28) MADs-box protein, GDEF1, partial
(58%)
Length = 1100
Score = 109 bits (273), Expect = 4e-25
Identities = 69/176 (39%), Positives = 103/176 (58%), Gaps = 6/176 (3%)
Frame = +2
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANN 77
MGRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+LI+FS +++EY +
Sbjct: 137 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYISP 316
Query: 78 --SVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEAL-- 133
+ K I++Y+K D S + + + KL+ + L+ R LGE L
Sbjct: 317 GLTTKRIIDQYQKTLGDIDLWRS----HYEKMLENLKKLKEINNKLRRQIRHRLGEGLDM 484
Query: 134 SNMNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQK--REIDLHNSNQLLRAK 187
+++ + L+ LE + I +IR +K ++ + +K R ++ N N LL K
Sbjct: 485 DDLSFQQLRKLEEDMVSSIGKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELK 652
>TC17763 similar to UP|Q7Y1U9 (Q7Y1U9) SVP-like floral repressor, partial
(35%)
Length = 737
Score = 93.2 bits (230), Expect = 4e-20
Identities = 45/70 (64%), Positives = 58/70 (82%)
Frame = +3
Query: 17 KMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYAN 76
+M R KI+IK+I+N T RQVTF KRR GL KKA ELSVLCDA+VAL+VFS+ G+L+EY+N
Sbjct: 498 EMAREKIQIKKIDNATARQVTFSKRRRGLFKKAEELSVLCDADVALVVFSSTGKLFEYSN 677
Query: 77 NSVKASIERY 86
S+K +ER+
Sbjct: 678 LSMKEILERH 707
>TC15233 similar to UP|AAS45681 (AAS45681) AGAMOUS-like protein (Fragment),
partial (40%)
Length = 610
Score = 87.8 bits (216), Expect = 2e-18
Identities = 48/112 (42%), Positives = 68/112 (59%)
Frame = +3
Query: 136 MNARDLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERK 195
++ ++LKNLE +LEKG+SR+RS+K+E LFA++E+MQKREI+L N N LRAKIAE + +
Sbjct: 3 LSLKELKNLEGRLEKGLSRVRSRKHETLFADVEFMQKREIELQNHNHYLRAKIAEHERAQ 182
Query: 196 NHNFNMLPGTTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
N + R FF L ++NQ +RQDQ +L V
Sbjct: 183 QQQQQQQQQPQNLMLSESLPSQSYDRNFFP-ANLLGSDNQYSRQDQTALPLV 335
>AV421179
Length = 207
Score = 83.6 bits (205), Expect = 3e-17
Identities = 40/67 (59%), Positives = 54/67 (79%)
Frame = +3
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANN 77
M R +I+IK+I+N ++RQVTF KRR GL KKA ELS LCDA++ALIVFS +L+EYA++
Sbjct: 6 MTRKRIQIKKIDNISSRQVTFSKRRKGLFKKAQELSTLCDADIALIVFSATNKLFEYASS 185
Query: 78 SVKASIE 84
S++ IE
Sbjct: 186SIQKVIE 206
>TC11647 similar to UP|Q7X9H7 (Q7X9H7) MADS-box protein AGL62, partial (25%)
Length = 843
Score = 82.8 bits (203), Expect = 5e-17
Identities = 48/121 (39%), Positives = 72/121 (58%), Gaps = 3/121 (2%)
Frame = +1
Query: 6 QSMSANNSPQRKMGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVF 65
Q + + S ++ +GR KIE+K++ N +N QVTF KRR+GL KKA EL +LCD E+AL+VF
Sbjct: 49 QPATMSTSRRKSLGRQKIEMKKMTNESNLQVTFSKRRSGLFKKASELCILCDVEIALVVF 228
Query: 66 SNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQFYQQEAAK---LRVQISNLQ 122
S +++ + + SV+A I+RY S TS Q +K L V++S +
Sbjct: 229 SPGEKVFSFGHPSVEAVIKRY-----FSQAPPQTSDTMQYIEAQRNSKVHELNVELSQIN 393
Query: 123 N 123
N
Sbjct: 394 N 396
>BP064158
Length = 394
Score = 80.9 bits (198), Expect = 2e-16
Identities = 39/60 (65%), Positives = 49/60 (81%)
Frame = +3
Query: 18 MGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANN 77
MGR K++ +++ENTTNRQVTF KRRNGL+KKAYELSVLC +VALI+FS GR ++ N
Sbjct: 165 MGRVKLQNQKVENTTNRQVTFSKRRNGLIKKAYELSVLCXFDVALIMFSPSGRACLFSGN 344
>TC17857 similar to UP|Q8VWZ3 (Q8VWZ3) C-type MADS box protein, partial
(29%)
Length = 532
Score = 79.0 bits (193), Expect = 7e-16
Identities = 47/103 (45%), Positives = 64/103 (61%)
Frame = +2
Query: 145 ETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHNFNMLPG 204
E++LEKG+SR+RS+K+E LF +IE+MQKREI+L N N LLRAKIA+ + + ++L G
Sbjct: 2 ESRLEKGLSRVRSRKHETLFGDIEFMQKREIELPNHNNLLRAKIAQHERAQPQEQSLLQG 181
Query: 205 TTNFESLQQSQQPFDSRGFFQVTGLQPNNNQCARQDQISLQFV 247
QS R FF V PN+ + QDQ +LQ V
Sbjct: 182 NPCVSIPSQSY----DRNFFPVN--VPNH*YSSCQDQSALQLV 292
>AU252010
Length = 328
Score = 75.5 bits (184), Expect = 8e-15
Identities = 40/114 (35%), Positives = 71/114 (62%), Gaps = 2/114 (1%)
Frame = +1
Query: 29 ENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSNRGRLYEYANNSVKA--SIERY 86
EN++NRQVT+ KR+NG+LKKA E++VLCDA+V+LI+F+ G++++Y + S +ERY
Sbjct: 1 ENSSNRQVTYSKRKNGILKKAKEITVLCDAQVSLIIFAASGKMHDYISPSTTLVDMLERY 180
Query: 87 KKACSDSSGGGSTSGANAQFYQQEAAKLRVQISNLQNHNRQMLGEALSNMNARD 140
K + G A + E +L+ + +Q R + G+ ++++N ++
Sbjct: 181 HK-----TSGKRLWDAKHENLNGEIERLKKENDGMQIELRHLKGDDINSLNYKE 327
>TC19685 similar to UP|Q8H6F8 (Q8H6F8) MADS box protein GHMADS-2, partial
(40%)
Length = 508
Score = 52.8 bits (125), Expect = 6e-08
Identities = 29/55 (52%), Positives = 36/55 (64%)
Frame = +3
Query: 150 KGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDERKNHNFNMLPG 204
+GI+ IRSK +EML AEIEY Q REI+L N N LR KI +D + NM+ G
Sbjct: 3 RGITSIRSKPHEMLLAEIEYFQTREIELENENLCLRTKI--TDVERIQPVNMVSG 161
>AU089354
Length = 178
Score = 44.7 bits (104), Expect = 2e-05
Identities = 21/47 (44%), Positives = 33/47 (69%), Gaps = 2/47 (4%)
Frame = +3
Query: 44 GLLKKAYELSVLCDAEVALIVFSNRGRLYEYANNSVKA--SIERYKK 88
G+LKKA E++VLC A+V+LI F+ G++++Y S+ +ERY K
Sbjct: 3 GILKKAIEITVLCXAQVSLIXFAASGKMHDYIRPSITXVEMLERYXK 143
>TC13748 similar to UP|Q39401 (Q39401) MADS5 protein, partial (28%)
Length = 624
Score = 44.7 bits (104), Expect = 2e-05
Identities = 21/51 (41%), Positives = 31/51 (60%)
Frame = +1
Query: 143 NLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDLHNSNQLLRAKIAESDE 193
NLE +L+ + IRS+KN+ LF I +QK++ L N LL KI E ++
Sbjct: 1 NLEQQLDSALKHIRSRKNQALFESISELQKKDKALQEQNNLLTKKIKEKEK 153
>AV423802
Length = 295
Score = 26.9 bits (58), Expect = 3.3
Identities = 16/38 (42%), Positives = 25/38 (65%)
Frame = +3
Query: 140 DLKNLETKLEKGISRIRSKKNEMLFAEIEYMQKREIDL 177
DLK+L+T+LEK +SR+ S+ + + E+ EIDL
Sbjct: 192 DLKSLDTQLEKHLSRVWSRSVDTKRPKEEW----EIDL 293
>AV778498
Length = 552
Score = 26.9 bits (58), Expect = 3.3
Identities = 18/76 (23%), Positives = 30/76 (38%)
Frame = +2
Query: 47 KKAYELSVLCDAEVALIVFSNRGRLYEYANNSVKASIERYKKACSDSSGGGSTSGANAQF 106
KK EL +L E +Y ++ I +K CSDS + S N +
Sbjct: 359 KKVSELELLLKTE-------------QYRIQELEQQISTLEKRCSDSEADANKSLENVSY 499
Query: 107 YQQEAAKLRVQISNLQ 122
E + ++S+L+
Sbjct: 500 LTSELEASQARVSSLE 547
>AV418326
Length = 263
Score = 26.6 bits (57), Expect = 4.3
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = -1
Query: 63 IVFSNRGRLYEYANNSVKASIERYKKAC 90
I++ NRG ++ Y+NN V +E K C
Sbjct: 206 IIYLNRGIIHAYSNNIVILRMESKKGCC 123
>BP060280
Length = 384
Score = 25.4 bits (54), Expect = 9.5
Identities = 9/16 (56%), Positives = 12/16 (74%)
Frame = +2
Query: 195 KNHNFNMLPGTTNFES 210
KNH+FN PG+ +F S
Sbjct: 179 KNHSFNFQPGSVHFXS 226
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.128 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,147,294
Number of Sequences: 28460
Number of extensions: 32333
Number of successful extensions: 153
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 151
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 151
length of query: 247
length of database: 4,897,600
effective HSP length: 88
effective length of query: 159
effective length of database: 2,393,120
effective search space: 380506080
effective search space used: 380506080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0404.13


