

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0404.11
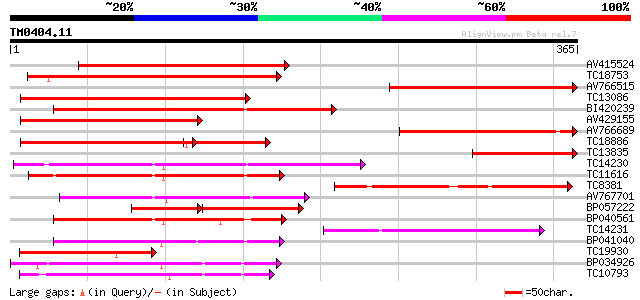
(365 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV415524 274 1e-74
TC18753 265 7e-72
AV766515 251 1e-67
TC13086 similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/hydrolas... 231 1e-61
BI420239 200 3e-52
AV429155 142 1e-34
AV766689 134 2e-32
TC18886 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/h... 127 2e-30
TC13835 126 5e-30
TC14230 126 7e-30
TC11616 similar to UP|Q9SVU5 (Q9SVU5) Proline-rich APG-like prot... 122 7e-29
TC8381 118 1e-27
AV767701 118 1e-27
BP057222 114 3e-26
BP040561 113 4e-26
TC14231 107 4e-24
BP041040 105 9e-24
TC19930 weakly similar to UP|Q9AX26 (Q9AX26) GDSL-motif lipase/h... 103 6e-23
BP034926 100 5e-22
TC10793 99 1e-21
>AV415524
Length = 411
Score = 274 bits (701), Expect = 1e-74
Identities = 135/136 (99%), Positives = 136/136 (99%)
Frame = +2
Query: 45 NNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDIL 104
+NNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDIL
Sbjct: 2 SNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDIL 181
Query: 105 RGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYS 164
RGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYS
Sbjct: 182 RGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYS 361
Query: 165 IGLGSNDYLNNYFMPQ 180
IGLGSNDYLNNYFMPQ
Sbjct: 362 IGLGSNDYLNNYFMPQ 409
>TC18753
Length = 562
Score = 265 bits (678), Expect = 7e-72
Identities = 128/167 (76%), Positives = 146/167 (86%), Gaps = 3/167 (1%)
Frame = +3
Query: 12 MLVMVLGLWGGW---VGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGP 68
+LV+++ LW VGA PQVPC+F FGDSLVDNGNNN L SLA+A+Y PYGIDFPGGP
Sbjct: 60 VLVLIVCLWSTTTTGVGAEPQVPCFFSFGDSLVDNGNNNRLSSLAKANYRPYGIDFPGGP 239
Query: 69 SGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRI 128
+GRFSNGKT+VDVI ELLGF +IPPY +T G DILRGVN+ASAAAGIREETGQQLGGRI
Sbjct: 240 TGRFSNGKTSVDVIGELLGFGSYIPPYATTRGQDILRGVNYASAAAGIREETGQQLGGRI 419
Query: 129 SFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNN 175
SF GQVQNYQ TVSQ++N+LG+E+ AANYLSKCIYSIG+GSNDYLNN
Sbjct: 420 SFRGQVQNYQRTVSQLINLLGDENTAANYLSKCIYSIGIGSNDYLNN 560
>AV766515
Length = 572
Score = 251 bits (641), Expect = 1e-67
Identities = 120/121 (99%), Positives = 120/121 (99%)
Frame = -3
Query: 245 INDANQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVG 304
INDA QIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVG
Sbjct: 570 INDATQIFNNKLKSVVDQFNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVG 391
Query: 305 RNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQ 364
RNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQ
Sbjct: 390 RNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQ 211
Query: 365 I 365
I
Sbjct: 210 I 208
>TC13086 similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif lipase/hydrolase-like
protein, partial (35%)
Length = 524
Score = 231 bits (590), Expect = 1e-61
Identities = 113/149 (75%), Positives = 130/149 (86%), Gaps = 1/149 (0%)
Frame = +3
Query: 8 MVVAMLVMVLGLWGGW-VGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
+V ++V+ LGLW VGA PQVPCYFIFGDSLVDNGNNN L S A+A+Y+PYGIDF G
Sbjct: 78 LVAVVMVVCLGLWSTTRVGADPQVPCYFIFGDSLVDNGNNNQLNS*AKANYLPYGIDFGG 257
Query: 67 GPSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGG 126
GP+GRFSNGKTTVDV+AELLGFD +IPPY + G DIL+GVN+ASAAAGIREETGQQLGG
Sbjct: 258 GPTGRFSNGKTTVDVVAELLGFDSYIPPYSTARGQDILKGVNYASAAAGIREETGQQLGG 437
Query: 127 RISFSGQVQNYQSTVSQVVNILGNEDQAA 155
RISFSGQV+NYQ TVSQVVN+LG+E+ AA
Sbjct: 438 RISFSGQVENYQRTVSQVVNLLGDENTAA 524
>BI420239
Length = 621
Score = 200 bits (509), Expect = 3e-52
Identities = 95/182 (52%), Positives = 129/182 (70%)
Frame = +1
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGGPSGRFSNGKTTVDVIAELLGF 88
QVPC FIFGDSL D+GNNN L + A+ DY+PYGIDFP GP+GR++NG+ +D I ELLG
Sbjct: 79 QVPCLFIFGDSLSDSGNNNNLETDAKVDYLPYGIDFPTGPTGRYTNGRNAIDKITELLGL 258
Query: 89 DDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNIL 148
+DFIPP+ + SG DIL+GVN+AS +AGIR+E+G LG I+ Q+ + + VSQ+ L
Sbjct: 259 EDFIPPFANLSGSDILKGVNYASGSAGIRKESGTNLGTNINMGLQIYFHMAIVSQISARL 438
Query: 149 GNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQYADVLIQAYTEQLQTL 208
G +A +LSKC+Y + +G+NDY NYF+P + TS +YT ++YA LI + LQ L
Sbjct: 439 GFH-KAKRHLSKCLYYVNIGTNDYEQNYFLPDLFDTSSKYTPEEYAKDLINRLSHYLQIL 615
Query: 209 YN 210
N
Sbjct: 616 RN 621
>AV429155
Length = 393
Score = 142 bits (357), Expect = 1e-34
Identities = 66/117 (56%), Positives = 89/117 (75%)
Frame = +3
Query: 8 MVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG 67
+ + +L++V V P+VPC FIFGDSL DNGNNN L + +++Y PYGIDFP G
Sbjct: 42 LFLPLLLLVANCMQHSVHGKPEVPCLFIFGDSLSDNGNNNNLVTSTKSNYKPYGIDFPTG 221
Query: 68 PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQL 124
P+GRF+NG TTVD+I++LLGF++FIPP+ +TS DIL+GVN+ASAAAGI E+G +
Sbjct: 222 PTGRFTNGPTTVDIISQLLGFENFIPPFANTSDSDILKGVNYASAAAGICVESGTHM 392
>AV766689
Length = 512
Score = 134 bits (338), Expect = 2e-32
Identities = 61/115 (53%), Positives = 85/115 (73%), Gaps = 1/115 (0%)
Frame = -1
Query: 252 FNNKLKSVVDQFNN-QLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQI 310
FN LK +V FN QLP A+ +Y++ Y QD+ + ++GF ++GCCGVGRNNGQI
Sbjct: 512 FNTGLKKMVQNFNGGQLPGAKFVYLDFYKSSQDLSTNGTSFGFEVVDKGCCGVGRNNGQI 333
Query: 311 TCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRLAQI 365
TCLP+Q PCENR +YLFWDAFHP+E N+++A+ YS+QS + YPI+I++LA +
Sbjct: 332 TCLPLQQPCENREKYLFWDAFHPTELANILLAKATYSSQSYT--YPINIQQLAML 174
>TC18886 weakly similar to UP|Q8LDT8 (Q8LDT8) GDSL-motif
lipase/hydrolase-like protein, partial (23%)
Length = 547
Score = 127 bits (320), Expect = 2e-30
Identities = 58/118 (49%), Positives = 89/118 (75%), Gaps = 4/118 (3%)
Frame = +1
Query: 8 MVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG 67
+V+++++MV + +G + VPC F+FGDSL D+GNNN L +L++A+++PYGIDFP G
Sbjct: 64 LVLSLVLMVACMQHSVLGNSQAVPCLFVFGDSLADSGNNNNLPTLSKANFLPYGIDFPTG 243
Query: 68 PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGDDILRGVNFASA----AAGIREETG 121
P+GR++NG +D +A++LGF+ FIPP+ + SG DIL+GVN+ASA +AG R + G
Sbjct: 244 PTGRYTNGLNPIDKLAQILGFEKFIPPFANLSGSDILKGVNYASAFSRNSAGNRNQFG 417
Score = 52.8 bits (125), Expect = 9e-08
Identities = 23/56 (41%), Positives = 39/56 (69%)
Frame = +2
Query: 113 AAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLG 168
+AGIR+ETG LG ++ Q+Q++++ VSQ+ LG +A NYL++C+Y + +G
Sbjct: 380 SAGIRQETGTNLGTNVNMGLQLQHHRTIVSQISTKLGGFHKAVNYLTQCLYYVYIG 547
>TC13835
Length = 450
Score = 126 bits (317), Expect = 5e-30
Identities = 53/67 (79%), Positives = 61/67 (90%)
Frame = -1
Query: 299 GCCGVGRNNGQITCLPMQTPCENRREYLFWDAFHPSEAGNVVIAQRAYSAQSPSDAYPID 358
GCCG+GRNNGQITCLP+Q PC NRREYLFWDAFHP+EAGN +I +RAY++QS SDAYPID
Sbjct: 450 GCCGIGRNNGQITCLPLQAPCSNRREYLFWDAFHPTEAGNTIIGRRAYNSQSASDAYPID 271
Query: 359 IKRLAQI 365
I RLAQ+
Sbjct: 270 INRLAQL 250
>TC14230
Length = 740
Score = 126 bits (316), Expect = 7e-30
Identities = 86/232 (37%), Positives = 127/232 (54%), Gaps = 5/232 (2%)
Frame = +1
Query: 3 LNVNMMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGI 62
L + LV+VL L +A +F+FGDSLVD+GNN+ L + ARAD PYGI
Sbjct: 58 LGFGFCTITSLVLVLFLSSV---SAQHTRAFFVFGDSLVDSGNNDFLATTARADAPPYGI 228
Query: 63 DFP-GGPSGRFSNGKTTVDVIAELLGFDDFIPPYVS--TSGDDILRGVNFASAAAGIREE 119
DFP P+GRFSNG D+I+ LG + + PY+S G+ +L G NFASA GI +
Sbjct: 229 DFPTHRPTGRFSNGLNIPDLISLELGLEPTL-PYLSPLLVGEKLLIGANFASAGIGILND 405
Query: 120 TGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMP 179
TG Q I Q++ +Q +V +G E N +++ ++ I LG ND++NNY++
Sbjct: 406 TGIQFLHIIHIEKQLKLFQQYQKRVSAHIGREG-TRNLVNRALFLITLGGNDFVNNYYLV 582
Query: 180 QFYSTSRQYTTDQYADVLIQAYTEQL-QTLYNFGARKMVLFGI-GQIGCSPN 229
+ + SRQY+ Y +I + + + + RK G+ GQ GC P+
Sbjct: 583 PYSARSRQYSLPDYVRYIISELPQSSEEDVMIWELRKGSGNGVQGQWGCVPS 738
>TC11616 similar to UP|Q9SVU5 (Q9SVU5) Proline-rich APG-like protein,
partial (41%)
Length = 569
Score = 122 bits (307), Expect = 7e-29
Identities = 73/168 (43%), Positives = 103/168 (60%), Gaps = 3/168 (1%)
Frame = +2
Query: 13 LVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPGG-PSGR 71
+V+V+ L G+ GA +F+FGDSLVDNGNNN L + ARAD PYGID P G P+GR
Sbjct: 74 IVLVVALASGFRGAEA-ARAFFVFGDSLVDNGNNNYLATTARADSPPYGIDTPNGRPTGR 250
Query: 72 FSNGKTTVDVIAELLGFDDFIPPYVS--TSGDDILRGVNFASAAAGIREETGQQLGGRIS 129
FSNG+ D I+E LG + + PY+S +GD++L G NFASA GI +TG Q I
Sbjct: 251 FSNGRNIPDFISEQLGAEPTL-PYLSPELNGDNLLVGANFASAGIGILNDTGIQFINIIR 427
Query: 130 FSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYF 177
Q++ + +V ++G E + ++ + I LG ND++NNY+
Sbjct: 428 IFRQLEYFDEYQQRVSALIGPE-ETQRLVNGALVLITLGGNDFVNNYY 568
>TC8381
Length = 844
Score = 118 bits (296), Expect = 1e-27
Identities = 58/154 (37%), Positives = 97/154 (62%), Gaps = 1/154 (0%)
Frame = +3
Query: 210 NFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQFNNQ-LP 268
+FGARK ++ G+ ++GC P R + +C+ N A +FN++LK++VD++N++ LP
Sbjct: 3 HFGARKTIMVGMDRLGCIPKA---RLTNNGSCIEKENVAAFLFNDQLKALVDRYNHKILP 173
Query: 269 DARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENRREYLFW 328
D++ I+IN+ I D ++GF+ T+ CC + G CLP TPC+NR +Y FW
Sbjct: 174 DSKFIFINSTAIIHD-----QSHGFTITDAACCQLNTTRG--VCLPNLTPCQNRSQYKFW 332
Query: 329 DAFHPSEAGNVVIAQRAYSAQSPSDAYPIDIKRL 362
D H +EA N++ A +YS P+ A+P++I++L
Sbjct: 333 DGIHTTEAANILTATVSYSTSDPNIAHPMNIQKL 434
>AV767701
Length = 622
Score = 118 bits (296), Expect = 1e-27
Identities = 67/164 (40%), Positives = 99/164 (59%), Gaps = 3/164 (1%)
Frame = +3
Query: 33 YFIFGDSLVDNGNNNALRSLARADYMPYGIDFP-GGPSGRFSNGKTTVDVIAELLGFDDF 91
+F+FGDSLVD+GNNN L + ARAD PYGID+P P+GRFSNG D+I++ LG +
Sbjct: 135 FFVFGDSLVDSGNNNYLATTARADSPPYGIDYPTRRPTGRFSNGLNIPDLISQQLGAESV 314
Query: 92 IPPYVSTS--GDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILG 149
+ PY+S G+ +L G NFASA GI +TG Q I Q+ ++ ++ + +G
Sbjct: 315 L-PYLSPQLRGNKLLLGANFASAGIGILNDTGTQFLNIIRMYRQLDYFEEYQHRLASQIG 491
Query: 150 NEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYSTSRQYTTDQY 193
A + K + I +G ND++NNY++ + + SRQY+ Y
Sbjct: 492 VTKTKA-LVDKALVLITVGGNDFVNNYYLVPYSARSRQYSLPDY 620
>BP057222
Length = 531
Score = 114 bits (285), Expect = 3e-26
Identities = 52/65 (80%), Positives = 60/65 (92%)
Frame = +2
Query: 125 GGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFMPQFYST 184
GGRISF GQVQNYQ TVSQ++N+LG+E+ AANYLSKCIYSIG+GSNDYLNNYFMP YS+
Sbjct: 329 GGRISFRGQVQNYQRTVSQLINLLGDENTAANYLSKCIYSIGIGSNDYLNNYFMPLIYSS 508
Query: 185 SRQYT 189
SRQ+T
Sbjct: 509 SRQFT 523
Score = 73.2 bits (178), Expect = 7e-14
Identities = 34/46 (73%), Positives = 39/46 (83%)
Frame = +3
Query: 79 VDVIAELLGFDDFIPPYVSTSGDDILRGVNFASAAAGIREETGQQL 124
V++ ELLGF +IPPY +T G DILRGVN+ASAAAGIREETGQQL
Sbjct: 60 VNITGELLGFGSYIPPYATTRGQDILRGVNYASAAAGIREETGQQL 197
>BP040561
Length = 515
Score = 113 bits (283), Expect = 4e-26
Identities = 69/156 (44%), Positives = 94/156 (60%), Gaps = 6/156 (3%)
Frame = +1
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFP-GGPSGRFSNGKTTVDVIAELLG 87
Q +F+FGDSLVDNGNN+ L + ARAD PYGIDFP P+GRFSNG D+I+E LG
Sbjct: 61 QTRAFFVFGDSLVDNGNNDFLATTARADAPPYGIDFPTHKPTGRFSNGLNIPDLISEQLG 240
Query: 88 FDDFIPPYVS--TSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQV---QNYQSTVS 142
+ + PY+S G+ +L G NFASA GI +TG Q I Q+ Q+YQ +S
Sbjct: 241 LEPTM-PYLSPLLMGEKLLVGANFASAGIGILNDTGFQFLNIIHMYKQLKLFQHYQQRLS 417
Query: 143 QVVNILGNEDQAANYLSKCIYSIGLGSNDYLNNYFM 178
+ G + ++K + I LG ND++NNY++
Sbjct: 418 AQIGPEGTK----KLVNKALVLITLGGNDFVNNYYL 513
>TC14231
Length = 803
Score = 107 bits (266), Expect = 4e-24
Identities = 54/142 (38%), Positives = 79/142 (55%)
Frame = +2
Query: 203 EQLQTLYNFGARKMVLFGIGQIGCSPNELAQRSQDGRTCVSDINDANQIFNNKLKSVVDQ 262
E L+ +Y+ GAR++++ G G +GC P LA RS+ G C ++ A ++N +L +++
Sbjct: 8 EVLRKMYDLGARRVLVTGTGPMGCVPPXLAMRSRTG-DCDVELXRAASLYNPQLVNMIKG 184
Query: 263 FNNQLPDARVIYINAYGIFQDIIASPATYGFSNTNEGCCGVGRNNGQITCLPMQTPCENR 322
N +L I NAY + D I++P YGF + CCG G NG C P C NR
Sbjct: 185 LNQELGADVFIAANAYQMHMDFISNPRAYGFVTSKIACCGQGPYNGIGLCTPASNLCPNR 364
Query: 323 REYLFWDAFHPSEAGNVVIAQR 344
Y FWD FHPSE + +I Q+
Sbjct: 365 DIYAFWDPFHPSERASRIIVQQ 430
>BP041040
Length = 550
Score = 105 bits (263), Expect = 9e-24
Identities = 59/152 (38%), Positives = 90/152 (58%), Gaps = 3/152 (1%)
Frame = +3
Query: 29 QVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFP-GGPSGRFSNGKTTVDVIAELLG 87
+VP +FGDS VD+GNNN + ++A++++ PYG DFP G P+GRFSNG+ D I+E G
Sbjct: 75 KVPAIIVFGDSSVDSGNNNFIPTMAKSNFEPYGRDFPDGNPTGRFSNGRIAPDFISEAFG 254
Query: 88 FDDFIPPYV--STSGDDILRGVNFASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVV 145
IP Y+ + S D GV FASA G T + I +V+ Y+ ++V
Sbjct: 255 LKPTIPAYLDPAYSISDFASGVCFASAGTGYDNST-SNVADVIPLWKEVEYYKDYRQKLV 431
Query: 146 NILGNEDQAANYLSKCIYSIGLGSNDYLNNYF 177
LG+E +A + + +Y + +G+ND+L NY+
Sbjct: 432 AYLGDE-KANEIVKEALYLVSIGTNDFLENYY 524
>TC19930 weakly similar to UP|Q9AX26 (Q9AX26) GDSL-motif
lipase/hydrolase-like protein, partial (11%)
Length = 329
Score = 103 bits (256), Expect = 6e-23
Identities = 48/90 (53%), Positives = 66/90 (73%), Gaps = 2/90 (2%)
Frame = +2
Query: 7 MMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
++ + +L++ + + +V QVPC F+FGDSL D+GNNN L +LA+ Y PYGIDFP
Sbjct: 59 IIALVVLLVAIAIMQIFVQRNTQVPCLFVFGDSLSDSGNNNNLETLAKVAYPPYGIDFPT 238
Query: 67 G--PSGRFSNGKTTVDVIAELLGFDDFIPP 94
G P+GR+SNG+T VD + ELLGF+DFIPP
Sbjct: 239 GPTPTGRYSNGRTAVDKLTELLGFEDFIPP 328
>BP034926
Length = 607
Score = 100 bits (248), Expect = 5e-22
Identities = 62/186 (33%), Positives = 103/186 (55%), Gaps = 11/186 (5%)
Frame = +2
Query: 1 MDLNVNMMVVAMLVMV--------LGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSL 52
M+L V ++++A+ +M+ + L W A V C +FGDS VD GNNN LR+
Sbjct: 59 MELMVKVVLLALAIMMPWCSFAVDIQLARQWA-AKSNVSCILVFGDSSVDPGNNNVLRTS 235
Query: 53 ARADYMPYGIDFPGG-PSGRFSNGKTTVDVIAELLGFDDFIPPYV--STSGDDILRGVNF 109
++++ PYG DF P+GRF NG+ D IAE LG+ +P ++ + +D+ GV+F
Sbjct: 236 MKSNFPPYGKDFFNSLPTGRFCNGRLATDFIAEALGYRQMLPAFLDPNLKVEDLPYGVSF 415
Query: 110 ASAAAGIREETGQQLGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGS 169
ASAA G + T + + S Q+Q + + +LG E++A + ++ + +G+
Sbjct: 416 ASAATGFDDYTANVV-NVLPVSKQIQYFMHYKIHLRKLLG-EERAEFIIRNALFIVSMGT 589
Query: 170 NDYLNN 175
ND+L N
Sbjct: 590 NDFLQN 607
>TC10793
Length = 556
Score = 99.0 bits (245), Expect = 1e-21
Identities = 62/167 (37%), Positives = 95/167 (56%), Gaps = 3/167 (1%)
Frame = +1
Query: 7 MMVVAMLVMVLGLWGGWVGAAPQVPCYFIFGDSLVDNGNNNALRSLARADYMPYGIDFPG 66
++ +L+ +L L V A +VP +FGDS VD GNNN + ++AR+++ PYG DF G
Sbjct: 73 LLCTTLLLQLLCL----VVAGGKVPAIIVFGDSSVDAGNNNFIETVARSNFQPYGRDFQG 240
Query: 67 G-PSGRFSNGKTTVDVIAELLGFDDFIPPYVSTSGD--DILRGVNFASAAAGIREETGQQ 123
G P+GRFSNG+ D I+E G ++P Y+ S + GV FASAA G T
Sbjct: 241 GKPTGRFSNGRIATDFISEAFGIKPYVPAYLDPSYNISHFATGVAFASAATGYDNATSDV 420
Query: 124 LGGRISFSGQVQNYQSTVSQVVNILGNEDQAANYLSKCIYSIGLGSN 170
L I Q++ Y++ ++ LG E +A + ++K ++ I LG+N
Sbjct: 421 L-SVIPLWKQLEYYKAYQKKLSTYLG-EKKAHDTITKSLHIISLGTN 555
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,714,117
Number of Sequences: 28460
Number of extensions: 71533
Number of successful extensions: 492
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 439
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 439
length of query: 365
length of database: 4,897,600
effective HSP length: 91
effective length of query: 274
effective length of database: 2,307,740
effective search space: 632320760
effective search space used: 632320760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0404.11


