

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
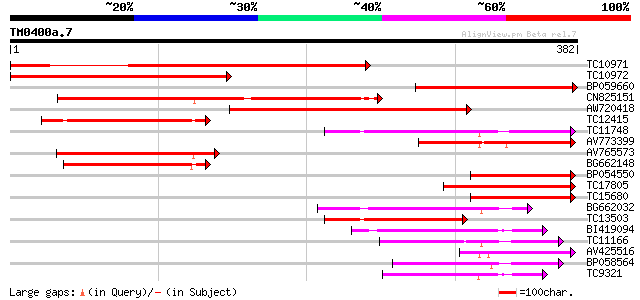
Query= TM0400a.7
(382 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10971 similar to UP|Q40318 (Q40318) Coil protein, partial (42%) 365 e-102
TC10972 similar to UP|Q40318 (Q40318) Coil protein, partial (32%) 303 2e-83
BP059660 237 3e-63
CN825151 212 7e-56
AW720418 197 2e-51
TC12415 similar to UP|Q9LII9 (Q9LII9) Genomic DNA, chromosome 3,... 137 3e-33
TC11748 weakly similar to UP|O82076 (O82076) Lipase homolog (Fra... 132 7e-32
AV773399 118 1e-27
AV765573 117 2e-27
BG662148 106 7e-24
BP054550 93 6e-20
TC17805 weakly similar to UP|O82681 (O82681) Lanatoside 15'-O-ac... 93 6e-20
TC15680 weakly similar to UP|Q7Y1X1 (Q7Y1X1) ENSP-like protein, ... 91 3e-19
BG662032 88 2e-18
TC13503 similar to UP|Q8W0Y7 (Q8W0Y7) Enod8.3 (Fragment), partia... 82 1e-16
BI419094 80 7e-16
TC11166 weakly similar to UP|Q9FXJ3 (Q9FXJ3) F1K23.17, partial (... 77 4e-15
AV425516 72 2e-13
BP058564 70 6e-13
TC9321 similar to UP|O04320 (O04320) Proline-rich protein APG is... 66 8e-12
>TC10971 similar to UP|Q40318 (Q40318) Coil protein, partial (42%)
Length = 615
Score = 365 bits (938), Expect = e-102
Identities = 189/243 (77%), Positives = 190/243 (77%)
Frame = +2
Query: 1 MNSRTLIHALWCFALCVACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNSDTGVVYAAFA 60
MNSRTLIHALWCFALCVACTFIQIPSG
Sbjct: 41 MNSRTLIHALWCFALCVACTFIQIPSG--------------------------------- 121
Query: 61 GLQSPGGISFFGNLSGRASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGAS 120
+GRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGAS
Sbjct: 122 -------------------NGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGAS 244
Query: 121 IRPVYGFSPFYLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTID 180
IRPVYGFSPFYLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTID
Sbjct: 245 IRPVYGFSPFYLGMQVAQFIQLQSHIENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTID 424
Query: 181 IGQNDLGFGLMHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSI 240
IGQNDLGFGLMHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNT PIGCLPTSSI
Sbjct: 425 IGQNDLGFGLMHTSEEEVLRSIPEMMRNFTYDVQVLYDVGARVFWIHNTCPIGCLPTSSI 604
Query: 241 FYE 243
FYE
Sbjct: 605 FYE 613
>TC10972 similar to UP|Q40318 (Q40318) Coil protein, partial (32%)
Length = 548
Score = 303 bits (777), Expect = 2e-83
Identities = 148/149 (99%), Positives = 149/149 (99%)
Frame = +1
Query: 1 MNSRTLIHALWCFALCVACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNSDTGVVYAAFA 60
MNSRTLIHALWCFALCVACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNS+TGVVYAAFA
Sbjct: 100 MNSRTLIHALWCFALCVACTFIQIPSGNGSYSSSSKCSYPAVYNFGDSNSNTGVVYAAFA 279
Query: 61 GLQSPGGISFFGNLSGRASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGAS 120
GLQSPGGISFFGNLSGRASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGAS
Sbjct: 280 GLQSPGGISFFGNLSGRASDGRLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGAS 459
Query: 121 IRPVYGFSPFYLGMQVAQFIQLQSHIENL 149
IRPVYGFSPFYLGMQVAQFIQLQSHIENL
Sbjct: 460 IRPVYGFSPFYLGMQVAQFIQLQSHIENL 546
>BP059660
Length = 501
Score = 237 bits (604), Expect = 3e-63
Identities = 109/109 (100%), Positives = 109/109 (100%)
Frame = -3
Query: 274 FQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGYRIDCGKKAVVNGT 333
FQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGYRIDCGKKAVVNGT
Sbjct: 499 FQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGSYYGYRIDCGKKAVVNGT 320
Query: 334 VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQACL 382
VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQACL
Sbjct: 319 VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQACL 173
>CN825151
Length = 740
Score = 212 bits (540), Expect = 7e-56
Identities = 113/225 (50%), Positives = 147/225 (65%), Gaps = 6/225 (2%)
Frame = +1
Query: 33 SSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEEL 92
+S C++PA+YNFGDSNSDTG + AAF + P G SF S R DGRLI+DFI E+L
Sbjct: 79 NSPPCAFPAIYNFGDSNSDTGGISAAFEPIPPPYGESFPQKPSARDCDGRLIVDFIAEKL 258
Query: 93 EIPYLSAYLNSIGSNYRHGANFAAGGASIRP------VYGFSPFYLGMQVAQFIQLQSHI 146
+PYLSAYLNS+G+NYRHGANFA GG++IR YG SPF L MQ QF Q ++
Sbjct: 259 NLPYLSAYLNSLGTNYRHGANFATGGSTIRRQNETIFQYGISPFSLDMQFVQFKQFKART 438
Query: 147 ENLLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRSIPEMM 206
+ L + + +S LP PE+FSKALYT DIGQNDL G + +++ S+P+++
Sbjct: 439 KQLYQEAKTALE----RSKLPVPEEFSKALYTFDIGQNDLSVGFRMMNFDQMRESMPDIV 606
Query: 207 RNFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDA 251
V+ +Y++G R FWIHNT PIGCLP ++FY K NL A
Sbjct: 607 NQLASAVKNIYELGGRTFWIHNTAPIGCLPV-NLFY---KHNLPA 729
>AW720418
Length = 500
Score = 197 bits (502), Expect = 2e-51
Identities = 100/164 (60%), Positives = 119/164 (71%), Gaps = 1/164 (0%)
Frame = +3
Query: 149 LLNQFSSNRTEPPFKSYLPRPEDFSKALYTIDIGQNDLGFGLMHTSEEEVLRSIPEMMRN 208
L Q N T F S +PR EDFS+ALY DIGQNDL +G ++SEE++ S+P ++
Sbjct: 9 LRTQILFNGTV*IFXSSIPRXEDFSRALYMFDIGQNDLSYGFQYSSEEQLRASLPNILSQ 188
Query: 209 FTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFY-EPKKGNLDANGCVIPHNKIAQEFNR 267
F+ V+ LY GARVFWIHNTGPIGCLP + + Y + KKGN+DANGCVI N IA EFN
Sbjct: 189 FSQAVEQLYXEGARVFWIHNTGPIGCLPLNFLAYGKSKKGNVDANGCVISQNGIAHEFNL 368
Query: 268 QLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLE 311
QLK+QV QLR+ LP AK YVDVY AKYEL+SNA K GFVNPLE
Sbjct: 369 QLKNQVLQLRKKLPLAKLIYVDVYKAKYELVSNARKLGFVNPLE 500
>TC12415 similar to UP|Q9LII9 (Q9LII9) Genomic DNA, chromosome 3, TAC
clone:K24A2, partial (22%)
Length = 366
Score = 137 bits (345), Expect = 3e-33
Identities = 70/114 (61%), Positives = 86/114 (75%)
Frame = +3
Query: 22 IQIPSGNGSYSSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDG 81
I I + S++ S+ YPA++NFGDSNSDTG + AAF + P G +F G LSGR SDG
Sbjct: 33 IVIVAAGVSFTEGSQ--YPAIFNFGDSNSDTGAISAAFTIVHPPNGQNFLGALSGRYSDG 206
Query: 82 RLIIDFITEELEIPYLSAYLNSIGSNYRHGANFAAGGASIRPVYGFSPFYLGMQ 135
RLIIDFITEEL++PYL AYLNS+G+NYRHGANFA GG+SI G+SPF+L Q
Sbjct: 207 RLIIDFITEELKLPYLDAYLNSVGANYRHGANFATGGSSILK-GGYSPFHLAYQ 365
>TC11748 weakly similar to UP|O82076 (O82076) Lipase homolog (Fragment),
partial (48%)
Length = 679
Score = 132 bits (333), Expect = 7e-32
Identities = 70/174 (40%), Positives = 96/174 (54%), Gaps = 5/174 (2%)
Frame = +3
Query: 213 VQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQ 272
V+ LY+ GAR FW+HNTGP+GCLP I +K +LD GC+ +N A+ FN L
Sbjct: 21 VKSLYNEGARKFWVHNTGPLGCLP--KILALAQKKDLDLFGCLSSYNSAARLFNEALYHS 194
Query: 273 VFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCG-----SYYGYRIDCGKK 327
+LR L A YVD+Y K++LI+NA+K GF NPL VCCG + R+ CG+
Sbjct: 195 SQKLRTRLKDATLVYVDIYAIKHDLIANATKYGFSNPLTVCCGFGGPPYNFDLRVTCGQP 374
Query: 328 AVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
C S++++WDG H+T+AAN ++A I S P P C
Sbjct: 375 GY-------QVCDEGSRYVNWDGTHHTEAANTFIASKILSTDYSTPRTPFEFFC 515
>AV773399
Length = 492
Score = 118 bits (296), Expect = 1e-27
Identities = 58/113 (51%), Positives = 71/113 (62%), Gaps = 7/113 (6%)
Frame = -1
Query: 276 LRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCG-----SYYGYRIDCGKKAVV 330
LR+ L A TYVDVY+ KY LIS A F PL CCG +YY I CG K V
Sbjct: 492 LRKKLHSASITYVDVYSVKYSLISQAKTHAFGEPLRACCGHGGKYNYY-IHIGCGAKLKV 316
Query: 331 NGT--VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
+G + G PC++PS ++WDGVH+T+AANKWV I DGS SDPP+P+ AC
Sbjct: 315 HGQEILLGKPCEDPSLSVNWDGVHFTEAANKWVFDQIVDGSFSDPPIPLNMAC 157
>AV765573
Length = 440
Score = 117 bits (294), Expect = 2e-27
Identities = 59/116 (50%), Positives = 79/116 (67%), Gaps = 6/116 (5%)
Frame = +2
Query: 32 SSSSKCSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEE 91
+++ C +PA++NFG SNSDTG AAF P G ++FG +GR SDGR+I+DFI +
Sbjct: 86 TTTKTCDFPAIFNFGASNSDTGGFAAAFLQPGPPQGDTYFGRPAGRFSDGRIILDFIAQN 265
Query: 92 LEIPYLSAYLNSIGSNYRHGANFAAGGASIR------PVYGFSPFYLGMQVAQFIQ 141
E+PYLSAYLNS+G+NY HGA+FA ++IR P SPF+LG+Q QF Q
Sbjct: 266 FELPYLSAYLNSLGANYSHGASFATLSSTIRLPESVVPNGRSSPFFLGLQYLQFQQ 433
>BG662148
Length = 415
Score = 106 bits (264), Expect = 7e-24
Identities = 55/105 (52%), Positives = 73/105 (69%), Gaps = 6/105 (5%)
Frame = +2
Query: 37 CSYPAVYNFGDSNSDTGVVYAAFAGLQSPGGISFFGNLSGRASDGRLIIDFITEELEIPY 96
C +PA++NFG SNSDTG AAF G + P G ++F +GR SDGR+IIDFI + + Y
Sbjct: 104 CDFPAIFNFGASNSDTGGYAAAFEGPKPPHGDTYFHRPAGRYSDGRIIIDFIAQSFGLTY 283
Query: 97 LSAYLNSIGSNYRHGANFAAGGASI------RPVYGFSPFYLGMQ 135
LSAYL+S+GSN+ HGANFA G++I +P SPF+LG+Q
Sbjct: 284 LSAYLDSLGSNFSHGANFATLGSTIILPKNVKP--RGSPFFLGVQ 412
>BP054550
Length = 417
Score = 93.2 bits (230), Expect = 6e-20
Identities = 38/73 (52%), Positives = 54/73 (73%), Gaps = 2/73 (2%)
Frame = -3
Query: 311 EVCCGSYY-GYRIDCGKKAVVNGT-VYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDG 368
E+CCG Y G+ ++CGKK ++NG +Y PC++PSQ+I WDGV Y +AAN+W+A I G
Sbjct: 415 EICCGYYKDGHPVNCGKKEIINGKEIYAGPCEDPSQYIRWDGVPYPEAANRWIAHRILHG 236
Query: 369 SLSDPPVPIGQAC 381
S SDPP+P+ +C
Sbjct: 235 SFSDPPLPLTHSC 197
>TC17805 weakly similar to UP|O82681 (O82681) Lanatoside
15'-O-acetylesterase precursor, partial (12%)
Length = 484
Score = 93.2 bits (230), Expect = 6e-20
Identities = 46/91 (50%), Positives = 58/91 (63%), Gaps = 2/91 (2%)
Frame = +3
Query: 293 AKYELISNASKQGFVNPLEVCCGSYYG-YRIDCGKKAVVNGT-VYGNPCKNPSQHISWDG 350
AKY LIS+ +GFV+PL +C G + I G NG V+GN C+ PS ++SWDG
Sbjct: 6 AKYGLISHTQHEGFVDPLPICFGYHVNDTHIWFGTIGTANGKDVFGNACEKPSMYVSWDG 185
Query: 351 VHYTQAANKWVAKHIRDGSLSDPPVPIGQAC 381
VHY +AAN WVA I +GS +DPP I QAC
Sbjct: 186 VHYAEAANHWVANRILNGSFTDPPTLITQAC 278
>TC15680 weakly similar to UP|Q7Y1X1 (Q7Y1X1) ENSP-like protein, partial
(7%)
Length = 641
Score = 90.9 bits (224), Expect = 3e-19
Identities = 40/72 (55%), Positives = 50/72 (68%), Gaps = 1/72 (1%)
Frame = +1
Query: 311 EVCCGSYYG-YRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGS 369
E CCG+Y G Y+I CG+ +GTV CKNPS+ +SWDGVHY++ AN +AK I G+
Sbjct: 1 EFCCGNYLGGYKISCGQNTTESGTVSDKTCKNPSEFLSWDGVHYSEEANLLIAKQILSGA 180
Query: 370 LSDPPVPIGQAC 381
SDPPV I QAC
Sbjct: 181 FSDPPVTIRQAC 216
>BG662032
Length = 421
Score = 88.2 bits (217), Expect = 2e-18
Identities = 51/147 (34%), Positives = 73/147 (48%), Gaps = 2/147 (1%)
Frame = +1
Query: 208 NFTYDVQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNR 267
+F + LY + AR F I N GPIGC+P Y+ L+ N CV NK+A ++N
Sbjct: 16 HFRAQLTRLYRLDARKFVIGNVGPIGCIP-----YQKTINQLNENECVDLPNKLALQYNA 180
Query: 268 QLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLEVCCGS--YYGYRIDCG 325
+LKD + +L NLP A F +VY ELI N K GF + CCG+ + + CG
Sbjct: 181 RLKDLMAELNDNLPGATFVVANVYDLVMELIKNYDKYGFTTSTKACCGNGGQFAGIVPCG 360
Query: 326 KKAVVNGTVYGNPCKNPSQHISWDGVH 352
+ + C + +H+ WD H
Sbjct: 361 PTSSI--------CTDRYKHVFWDPYH 417
>TC13503 similar to UP|Q8W0Y7 (Q8W0Y7) Enod8.3 (Fragment), partial (30%)
Length = 705
Score = 82.0 bits (201), Expect = 1e-16
Identities = 42/96 (43%), Positives = 59/96 (60%)
Frame = -2
Query: 213 VQVLYDVGARVFWIHNTGPIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQ 272
++++YD+ AR F IHNT PIGCLP +I K D GC +N++A+ FN++LK+
Sbjct: 392 IKLIYDLRARSFGIHNT*PIGCLP--AILTNFPKAERDHYGCAT*YNEVAKYFNQKLKEA 219
Query: 273 VFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVN 308
+ QLRR LP A YVD+Y+ K + N K G N
Sbjct: 218 LAQLRRELPLAAIIYVDIYSPKLDHFRNLEKYGETN 111
>BI419094
Length = 585
Score = 79.7 bits (195), Expect = 7e-16
Identities = 45/133 (33%), Positives = 70/133 (51%), Gaps = 1/133 (0%)
Frame = +3
Query: 231 PIGCLPTSSIFYEPKKGNLDANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDV 290
P+GCLP + + D+N CV N A FNR+L L+++LP K +D+
Sbjct: 15 PVGCLPAAITLF-----GHDSNQCVARLNNDAVNFNRKLNTTSQSLQKSLPGLKLVLLDI 179
Query: 291 YTAKYELISNASKQGFVNPLEVCCGS-YYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWD 349
Y Y+L++ S+ GF CCG+ I C +K++ GT C N S+++ WD
Sbjct: 180 YQPLYDLVTKPSENGFAEARRACCGTGLLETSILCNQKSI--GT-----CANASEYVFWD 338
Query: 350 GVHYTQAANKWVA 362
G H ++AAN+ +A
Sbjct: 339 GFHPSEAANQVLA 377
>TC11166 weakly similar to UP|Q9FXJ3 (Q9FXJ3) F1K23.17, partial (15%)
Length = 495
Score = 77.4 bits (189), Expect = 4e-15
Identities = 44/128 (34%), Positives = 65/128 (50%), Gaps = 4/128 (3%)
Frame = +2
Query: 250 DANGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNP 309
D GCV N+ A+ +N++L+ ++ +LR P A Y D Y A L N K GF
Sbjct: 2 DQAGCVKWLNEFAEFYNQKLQLEIHRLRELHPHANIIYADYYNAALPLYQNPQKFGF-RG 178
Query: 310 LEVCCGS----YYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHI 365
LE+CCG Y CG V+ C +PS++I WDG+H T+AA +++ +
Sbjct: 179 LEICCGMGGPYNYNASAGCGSPGVI-------ACDDPSEYIGWDGIHLTEAAYGFISDGL 337
Query: 366 RDGSLSDP 373
+G S P
Sbjct: 338 MNGPYSFP 361
>AV425516
Length = 415
Score = 72.0 bits (175), Expect = 2e-13
Identities = 36/83 (43%), Positives = 49/83 (58%), Gaps = 5/83 (6%)
Frame = -3
Query: 304 QGFVNPLEVCCG--SYYGYR--IDCGKKAVVNGT-VYGNPCKNPSQHISWDGVHYTQAAN 358
QGF PL CCG Y Y + CG+ V+G ++ C++PS + WDG HYT+AAN
Sbjct: 392 QGFEIPLINCCGYGGKYNYTDGVACGQNIKVDGEEIFVGSCESPSTKVIWDGTHYTEAAN 213
Query: 359 KWVAKHIRDGSLSDPPVPIGQAC 381
K V I G+ SDPP+P+ +C
Sbjct: 212 KIVFDLISTGAFSDPPIPLNMSC 144
>BP058564
Length = 468
Score = 70.1 bits (170), Expect = 6e-13
Identities = 39/120 (32%), Positives = 59/120 (48%), Gaps = 5/120 (4%)
Frame = -2
Query: 259 NKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFV-NPLEVCCGSY 317
N + + L+ ++ QLR P Y D + A ++ + GF N L+VCCG
Sbjct: 452 NMFHEHHDDLLQIELNQLRVLYPLTNIIYADFFNAALQIYKSPENFGFGGNALKVCCGGG 273
Query: 318 YGYRID----CGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVAKHIRDGSLSDP 373
Y + CG V+ C +PSQ++SWDG H T+AA +W+ K + DGS + P
Sbjct: 272 GPYNYNDTAVCGNSGVI-------ACDDPSQYVSWDGYHLTEAAYRWITKGLLDGSYTTP 114
>TC9321 similar to UP|O04320 (O04320) Proline-rich protein APG isolog,
partial (37%)
Length = 541
Score = 66.2 bits (160), Expect = 8e-12
Identities = 37/113 (32%), Positives = 57/113 (49%), Gaps = 2/113 (1%)
Frame = +2
Query: 252 NGCVIPHNKIAQEFNRQLKDQVFQLRRNLPKAKFTYVDVYTAKYELISNASKQGFVNPLE 311
NGCV N AQ FN+++ +L++ P K D+Y Y+L+ S GF
Sbjct: 5 NGCVSRFNTDAQAFNKKINSAAAKLQKQHPGLKIVIFDIYKPLYDLVQTPSNFGFAEARR 184
Query: 312 VCC--GSYYGYRIDCGKKAVVNGTVYGNPCKNPSQHISWDGVHYTQAANKWVA 362
CC G+ + C K++ GT C N +Q++ WD VH ++AAN+ +A
Sbjct: 185 GCCGTGTVETTSLLCNPKSI--GT-----CSNATQYVFWDSVHPSEAANQVLA 322
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,132,053
Number of Sequences: 28460
Number of extensions: 103808
Number of successful extensions: 658
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 623
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 627
length of query: 382
length of database: 4,897,600
effective HSP length: 92
effective length of query: 290
effective length of database: 2,279,280
effective search space: 660991200
effective search space used: 660991200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0400a.7


