

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
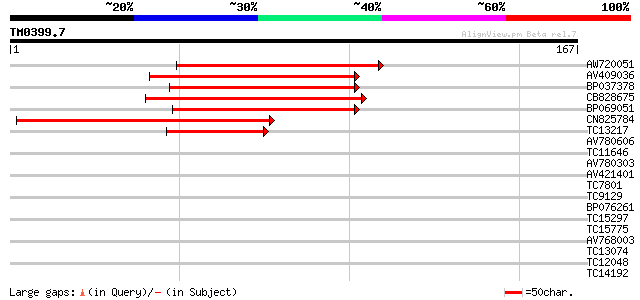
Query= TM0399.7
(167 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AW720051 100 1e-22
AV409036 99 4e-22
BP037378 98 6e-22
CB828675 91 8e-20
BP069051 86 2e-18
CN825784 71 8e-14
TC13217 similar to UP|Q9M4G1 (Q9M4G1) Dof zinc finger protein, p... 53 2e-08
AV780606 33 0.033
TC11646 27 1.8
AV780303 27 1.8
AV421401 27 1.8
TC7801 similar to UP|Q8VX01 (Q8VX01) Casein kinase II alpha subu... 27 2.3
TC9129 homologue to UP|Q8LLE2 (Q8LLE2) BEL1-related homeotic pro... 27 2.3
BP076261 26 3.0
TC15297 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zippe... 26 3.0
TC15775 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcri... 26 4.0
AV768003 26 4.0
TC13074 similar to GB|AAO42946.1|28416831|BT004700 At3g18880 {Ar... 26 4.0
TC12048 weakly similar to UP|Q9LGA3 (Q9LGA3) ESTs D22655(C0749),... 25 5.2
TC14192 similar to PIR|S29224|S29224 translation elongation fact... 25 5.2
>AW720051
Length = 564
Score = 100 bits (250), Expect = 1e-22
Identities = 39/61 (63%), Positives = 52/61 (84%)
Frame = +1
Query: 50 KIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRKMRPSCD 109
+I+ CPRC+S+ TKFCY+NNYN++QPRHFCKGC+RYWT GG LRN+ VG+G RK + S +
Sbjct: 130 QILKCPRCESLNTKFCYYNNYNLSQPRHFCKGCRRYWTKGGLLRNITVGSGSRKPKRSIN 309
Query: 110 E 110
+
Sbjct: 310 K 312
>AV409036
Length = 429
Score = 99.0 bits (245), Expect = 4e-22
Identities = 39/62 (62%), Positives = 49/62 (78%)
Frame = +1
Query: 42 RDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGR 101
R ++ ++ + CPRC S+ TKFCY+NNYN++QPRHFCK C+RYWT GG LRNVPVG G
Sbjct: 145 RPHHQQNNQALKCPRCDSINTKFCYYNNYNLSQPRHFCKNCRRYWTKGGVLRNVPVGGGC 324
Query: 102 RK 103
RK
Sbjct: 325 RK 330
>BP037378
Length = 546
Score = 98.2 bits (243), Expect = 6e-22
Identities = 39/56 (69%), Positives = 47/56 (83%)
Frame = +1
Query: 48 PDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
P + + CPRC+S TKFCYFNNY+++QPRHFCK C+RYWT GGALRNVPVG G R+
Sbjct: 223 PPEALKCPRCESTNTKFCYFNNYSLSQPRHFCKTCKRYWTRGGALRNVPVGGGFRR 390
>CB828675
Length = 552
Score = 91.3 bits (225), Expect = 8e-20
Identities = 34/65 (52%), Positives = 48/65 (73%)
Frame = +3
Query: 41 ERDQEKKPDKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAG 100
++ Q+ + + + CPRC S TKFCY+NNYN +QPRH+C+ C+R+WT GG LRNVPVG
Sbjct: 243 KQQQQSQQSEALKCPRCDSTNTKFCYYNNYNKSQPRHYCRACKRHWTKGGTLRNVPVGGV 422
Query: 101 RRKMR 105
R+ +
Sbjct: 423 RKNKK 437
>BP069051
Length = 359
Score = 86.3 bits (212), Expect = 2e-18
Identities = 32/55 (58%), Positives = 45/55 (81%)
Frame = -2
Query: 49 DKIIPCPRCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVPVGAGRRK 103
++ + CPRC S+ T+FCY+NNY++ QPR+FC C+RYWT GG+LR++PVG G RK
Sbjct: 304 EEAVNCPRCHSINTQFCYYNNYSLTQPRYFC*TCRRYWTEGGSLRHIPVGGGSRK 140
>CN825784
Length = 548
Score = 71.2 bits (173), Expect = 8e-14
Identities = 30/76 (39%), Positives = 46/76 (60%)
Frame = +1
Query: 3 QVQSGQDSEGIKLFGTTITLHGGRETRERSEDKKGSGEERDQEKKPDKIIPCPRCKSMET 62
+ ++ S G + T ++ + + +E+ KKPDK+IPCPRC SM+T
Sbjct: 319 ETKNSSTSPGAVVNPETPSIEEETAKSKSEQSDPAQSQEKTTLKKPDKVIPCPRCNSMDT 498
Query: 63 KFCYFNNYNVNQPRHF 78
KFCY++NY+VNQPR+F
Sbjct: 499 KFCYYHNYHVNQPRYF 546
>TC13217 similar to UP|Q9M4G1 (Q9M4G1) Dof zinc finger protein, partial
(15%)
Length = 422
Score = 53.1 bits (126), Expect = 2e-08
Identities = 19/30 (63%), Positives = 25/30 (83%)
Frame = +1
Query: 47 KPDKIIPCPRCKSMETKFCYFNNYNVNQPR 76
+PD + CPRC+S TKFCY+NNYN++QPR
Sbjct: 331 QPDATLKCPRCESTNTKFCYYNNYNLSQPR 420
>AV780606
Length = 585
Score = 32.7 bits (73), Expect = 0.033
Identities = 28/100 (28%), Positives = 40/100 (40%), Gaps = 5/100 (5%)
Frame = -1
Query: 23 HGGRETRERSEDKKGSGEERDQEKKPDKI---IPCPRCKSMETKFCYFNNYNVNQPRHFC 79
H G+ T + +D +G DQE D C C + +K + PR C
Sbjct: 534 HKGQSTSSKKQDG-ANGSGTDQESGQDDSQAETSCTHCGT-SSKSTPMMRRGPSGPRSLC 361
Query: 80 KGCQRYWTAGGALRNVPVGAGRRKMRPSC--DELLDGGGS 117
C +W GALR++ +R S E +DGG S
Sbjct: 360 NACGLFWANRGALRDL----SKRNQEHSLVQVEQVDGGNS 253
>TC11646
Length = 519
Score = 26.9 bits (58), Expect = 1.8
Identities = 12/30 (40%), Positives = 14/30 (46%), Gaps = 2/30 (6%)
Frame = +3
Query: 53 PC--PRCKSMETKFCYFNNYNVNQPRHFCK 80
PC P C S + K C+ NY HF K
Sbjct: 15 PCFVPECLSSQLKTCFIMNYKGASELHFAK 104
>AV780303
Length = 562
Score = 26.9 bits (58), Expect = 1.8
Identities = 10/27 (37%), Positives = 18/27 (66%)
Frame = +3
Query: 24 GGRETRERSEDKKGSGEERDQEKKPDK 50
GGR +R+R D + S +RD+++ +K
Sbjct: 45 GGRSSRDRDRDGERSSRDRDRDRGREK 125
>AV421401
Length = 419
Score = 26.9 bits (58), Expect = 1.8
Identities = 21/79 (26%), Positives = 28/79 (34%), Gaps = 3/79 (3%)
Frame = +3
Query: 89 GGALRNVPVGAGRRKMRPSCDELLDGGGSICDA---SATVEQLGLGTHREWHEAAAMFHG 145
G + R + G GRR +P C + +G C A E LG E + HG
Sbjct: 15 GSSGRCISHGGGRRCQKPGCHKGAEGRTVYCKAHGGGRRCEYLGCTKSAEGRTDYCIAHG 194
Query: 146 DFRQLFPSKRPRTTAGSGG 164
R+ R G G
Sbjct: 195 GGRRCSHEGCTRAARGKSG 251
>TC7801 similar to UP|Q8VX01 (Q8VX01) Casein kinase II alpha subunit
precursor , partial (28%)
Length = 591
Score = 26.6 bits (57), Expect = 2.3
Identities = 13/40 (32%), Positives = 20/40 (49%)
Frame = -1
Query: 17 GTTITLHGGRETRERSEDKKGSGEERDQEKKPDKIIPCPR 56
G G R R K+GSGE++ +E + ++PC R
Sbjct: 120 GEVAAEQGWRRRRSGGGRKRGSGEDKKEEN--EIVVPCGR 7
>TC9129 homologue to UP|Q8LLE2 (Q8LLE2) BEL1-related homeotic protein 13
(Fragment), partial (22%)
Length = 604
Score = 26.6 bits (57), Expect = 2.3
Identities = 14/40 (35%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Frame = +1
Query: 12 GIKLF----GTTITLHGGRETRERSEDKKGSGEERDQEKK 47
GIKL GT + HGGR+ R++ + ER + K+
Sbjct: 283 GIKLVHKCQGTVVETHGGRDVPTRTQRSRVCRRERKEPKQ 402
>BP076261
Length = 389
Score = 26.2 bits (56), Expect = 3.0
Identities = 9/19 (47%), Positives = 16/19 (83%)
Frame = +2
Query: 27 ETRERSEDKKGSGEERDQE 45
+ +R +D++GSGEER++E
Sbjct: 50 DKEDRRDDEEGSGEERERE 106
>TC15297 similar to UP|HT22_ARATH (P46604) Homeobox-leucine zipper protein
HAT22 (HD-ZIP protein 22), partial (32%)
Length = 585
Score = 26.2 bits (56), Expect = 3.0
Identities = 11/24 (45%), Positives = 15/24 (61%)
Frame = +2
Query: 95 VPVGAGRRKMRPSCDELLDGGGSI 118
+P+ A M PSC+ L DGG +I
Sbjct: 146 MPMSAATLTMCPSCERLGDGGSNI 217
>TC15775 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcription
factor 17 (WRKY DNA-binding protein 17), partial (53%)
Length = 1284
Score = 25.8 bits (55), Expect = 4.0
Identities = 14/42 (33%), Positives = 19/42 (44%), Gaps = 2/42 (4%)
Frame = -1
Query: 31 RSEDKKGSGEERDQEKKPDKIIPCPRCKSMET--KFCYFNNY 70
RSE + Q PD +P PR S +T + C NN+
Sbjct: 924 RSESHDRAHPRTSQSASPDHPLPAPRVSSPDTLSRCCTCNNH 799
>AV768003
Length = 362
Score = 25.8 bits (55), Expect = 4.0
Identities = 8/14 (57%), Positives = 10/14 (71%)
Frame = -2
Query: 70 YNVNQPRHFCKGCQ 83
++ QPRH C GCQ
Sbjct: 214 FHHKQPRHLCSGCQ 173
>TC13074 similar to GB|AAO42946.1|28416831|BT004700 At3g18880 {Arabidopsis
thaliana;}, partial (79%)
Length = 673
Score = 25.8 bits (55), Expect = 4.0
Identities = 11/25 (44%), Positives = 13/25 (52%)
Frame = -2
Query: 16 FGTTITLHGGRETRERSEDKKGSGE 40
F T T HG R RER D++ E
Sbjct: 108 FRRTTTTHGNRRRRERERDERHRSE 34
>TC12048 weakly similar to UP|Q9LGA3 (Q9LGA3) ESTs D22655(C0749), partial
(20%)
Length = 697
Score = 25.4 bits (54), Expect = 5.2
Identities = 16/41 (39%), Positives = 19/41 (46%)
Frame = +1
Query: 56 RCKSMETKFCYFNNYNVNQPRHFCKGCQRYWTAGGALRNVP 96
R K KF +F+N +PR QR W A G LR P
Sbjct: 19 REKENSRKFTHFSNGG--KPRRRSIRIQRPWLAEGDLRETP 135
>TC14192 similar to PIR|S29224|S29224 translation elongation factor eEF-1
beta'' chain - rice {Oryza sativa;}, partial (91%)
Length = 1066
Score = 25.4 bits (54), Expect = 5.2
Identities = 16/47 (34%), Positives = 23/47 (48%)
Frame = +1
Query: 4 VQSGQDSEGIKLFGTTITLHGGRETRERSEDKKGSGEERDQEKKPDK 50
+ + +D + + LFG E EDKK + EER+ KKP K
Sbjct: 361 LDTAEDDDDLDLFGD-----------ETEEDKKAA-EEREAAKKPAK 465
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,375,785
Number of Sequences: 28460
Number of extensions: 54397
Number of successful extensions: 293
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 293
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 293
length of query: 167
length of database: 4,897,600
effective HSP length: 84
effective length of query: 83
effective length of database: 2,506,960
effective search space: 208077680
effective search space used: 208077680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0399.7


