

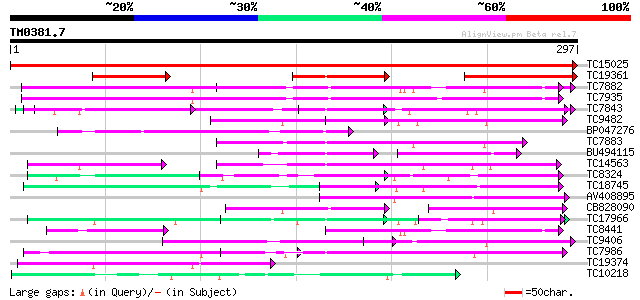
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0381.7
(297 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15025 similar to UP|MCAT_ARATH (Q93XM7) Mitochondrial carnitin... 516 e-147
TC19361 similar to GB|BAB08924.1|9758516|AB016882 carnitine/acyl... 122 7e-29
TC7882 homologue to UP|O49875 (O49875) Adenine nucleotide transl... 80 5e-16
TC7935 similar to UP|O49447 (O49447) ADP, ATP carrier-like prote... 77 3e-15
TC7843 weakly similar to GB|AAF03412.1|6090963|AF188712 mitochon... 71 3e-13
TC9482 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier prote... 69 1e-12
BP047276 62 9e-11
TC7883 homologue to UP|ADT1_GOSHI (O22342) ADP,ATP carrier prote... 62 1e-10
BU494115 40 1e-09
TC14563 similar to UP|Q9SSP4 (Q9SSP4) F3N23.2 protein (At1g72820... 58 2e-09
TC8324 UP|Q8VWY1 (Q8VWY1) Mitochondrial phosphate transporter, c... 58 2e-09
TC18745 weakly similar to UP|ADT2_HUMAN (P05141) ADP,ATP carrier... 57 4e-09
AV408895 55 2e-08
CB828090 53 5e-08
TC17966 similar to UP|Q8L9F8 (Q8L9F8) Mitochondrial carrier-like... 53 7e-08
TC8441 homologue to UP|ADT2_SOLTU (P27081) ADP,ATP carrier prote... 53 7e-08
TC9406 similar to UP|Q9M058 (Q9M058) Ca-dependent solute carrier... 52 9e-08
TC7986 similar to UP|Q8SF02 (Q8SF02) Dicarboxylate/tricarboxylat... 52 1e-07
TC19374 similar to UP|Q8LNZ1 (Q8LNZ1) Mitochondrial uncoupling p... 52 2e-07
TC10218 similar to UP|Q9SJ70 (Q9SJ70) Probable calcium-binding m... 52 2e-07
>TC15025 similar to UP|MCAT_ARATH (Q93XM7) Mitochondrial
carnitine/acylcarnitine carrier-like protein (A BOUT DE
SOUFFLE) (Carnitine/acylcarnitine translocase-like
protein) (CAC-like protein), partial (97%)
Length = 1256
Score = 516 bits (1328), Expect = e-147
Identities = 256/297 (86%), Positives = 270/297 (90%)
Frame = +2
Query: 1 MGDVAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAE 60
MGDVAKDL AGT GGA+QLIVGHPFDTIKVKLQSQPTP+PGQ P++SGA DAVKQT+AAE
Sbjct: 161 MGDVAKDLAAGTVGGASQLIVGHPFDTIKVKLQSQPTPLPGQPPKFSGAFDAVKQTIAAE 340
Query: 61 GPRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSF 120
GPRGL+KGMGAPLATVAA NAVLFTVRGQME + RSHPGA LTV QQ CGAGAG+AVSF
Sbjct: 341 GPRGLFKGMGAPLATVAAFNAVLFTVRGQMETIVRSHPGAPLTVSQQFICGAGAGVAVSF 520
Query: 121 LVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAR 180
L CPTELIKCRLQAQS L GSG AAVAVKYGGPMDVAR VLRSEGG RGLFKGLVPTMAR
Sbjct: 521 LACPTELIKCRLQAQSALGGSGAAAVAVKYGGPMDVARQVLRSEGGTRGLFKGLVPTMAR 700
Query: 181 EVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQV 240
E+PGNA MFG YEA KQ +AGG DTSGLGRGSLIVAGGLAGASFWF VYPTDVVKSVIQV
Sbjct: 701 EIPGNAIMFGVYEAIKQQIAGGTDTSGLGRGSLIVAGGLAGASFWFLVYPTDVVKSVIQV 880
Query: 241 DDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSALG 297
DDYK PKFSGS+DAFR+I A+EG KGLYKGFGPAM RS+PANAACFLAYEMTRSALG
Sbjct: 881 DDYKNPKFSGSLDAFRKIQATEGFKGLYKGFGPAMARSIPANAACFLAYEMTRSALG 1051
>TC19361 similar to GB|BAB08924.1|9758516|AB016882 carnitine/acylcarnitine
translocase-like protein {Arabidopsis thaliana;} ,
partial (20%)
Length = 515
Score = 122 bits (306), Expect = 7e-29
Identities = 59/59 (100%), Positives = 59/59 (100%)
Frame = +2
Query: 239 QVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSALG 297
QVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSALG
Sbjct: 2 QVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSALG 178
Score = 49.3 bits (116), Expect = 8e-07
Identities = 25/51 (49%), Positives = 32/51 (62%)
Frame = +2
Query: 149 KYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLL 199
K+ G +D R +L SEG +GL+KG P M R VP NAA F AYE ++ L
Sbjct: 26 KFSGSIDAFRRILASEG-IKGLYKGFGPAMFRSVPANAACFLAYEMTRSAL 175
Score = 42.7 bits (99), Expect = 7e-05
Identities = 19/41 (46%), Positives = 27/41 (65%)
Frame = +2
Query: 44 PRYSGAIDAVKQTLAAEGPRGLYKGMGAPLATVAALNAVLF 84
P++SG+IDA ++ LA+EG +GLYKG G + NA F
Sbjct: 23 PKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACF 145
>TC7882 homologue to UP|O49875 (O49875) Adenine nucleotide translocator,
complete
Length = 1656
Score = 79.7 bits (195), Expect = 5e-16
Identities = 81/294 (27%), Positives = 126/294 (42%), Gaps = 10/294 (3%)
Frame = +1
Query: 7 DLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIP-GQLPR-YSGAIDAVKQTLAAEGPRG 64
D G A P + +K+ +Q+Q I G+L Y G D +T EG
Sbjct: 463 DFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKTGRLSEPYKGIGDCFARTTKDEGIVA 642
Query: 65 LYKGMGAPLATVAALNAVLFTVRGQMEALF---RSHPGASLTVHQQVFCGAGAGLAVSFL 121
L++G A + A+ F + + LF + G + G AG +
Sbjct: 643 LWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAAGASSLLF 822
Query: 122 VCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMARE 181
V + + RL + A G ++ G +DV R L S+G A GL++G +
Sbjct: 823 VYSLDYARTRLANDAKAAKKGGER---QFNGLVDVYRKTLASDGVA-GLYRGFNISCVGI 990
Query: 182 VPGNAAMFGAYEASKQLLAGGP--DT--SGLGRGSLIVAG-GLAGASFWFFVYPTDVVKS 236
+ FG Y++ K +L G D+ + G G LI G GLA YP D V+
Sbjct: 991 IVYRGLYFGMYDSLKPVLLTGKLQDSFFASFGLGWLITNGAGLAS-------YPIDTVRR 1149
Query: 237 VIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYE 290
+ + + K+ S+DAF++IL +EG K L+KG G + R+V A A Y+
Sbjct: 1150RMMMTSGEAVKYKSSLDAFQQILKNEGAKSLFKGAGANILRAV-AGAGVLAGYD 1308
Score = 62.4 bits (150), Expect = 9e-11
Identities = 54/196 (27%), Positives = 83/196 (41%), Gaps = 8/196 (4%)
Frame = +1
Query: 109 FCGAGAGLAVS-FLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGA 167
F G AVS P E +K +Q Q + +G ++ Y G D + EG
Sbjct: 466 FLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKTGR--LSEPYKGIGDCFARTTKDEGIV 639
Query: 168 RGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGR--GSLIVAGGLAGASFW 225
L++G + R P A F + K+L D G + + +GG AGAS
Sbjct: 640 -ALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAAGASSL 816
Query: 226 FFVYPTDVVKSVIQVDDYKYPK-----FSGSIDAFRRILASEGIKGLYKGFGPAMFRSVP 280
FVY D ++ + D K F+G +D +R+ LAS+G+ GLY+GF + +
Sbjct: 817 LFVYSLDYARTRLANDAKAAKKGGERQFNGLVDVYRKTLASDGVAGLYRGFNISCVGIIV 996
Query: 281 ANAACFLAYEMTRSAL 296
F Y+ + L
Sbjct: 997 YRGLYFGMYDSLKPVL 1044
>TC7935 similar to UP|O49447 (O49447) ADP, ATP carrier-like protein,
partial (83%)
Length = 1465
Score = 77.0 bits (188), Expect = 3e-15
Identities = 75/289 (25%), Positives = 120/289 (40%), Gaps = 5/289 (1%)
Frame = +3
Query: 7 DLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIP-GQLPR-YSGAIDAVKQTLAAEGPRG 64
D G A P + IK+ +Q+Q I G+L Y G D +T EG
Sbjct: 252 DFLMGGVSAAVSKTAAAPIERIKLLIQNQDEMIKSGRLSEPYKGIGDCFARTTKDEGAIA 431
Query: 65 LYKGMGAPLATVAALNAVLFTVRGQMEALF---RSHPGASLTVHQQVFCGAGAGLAVSFL 121
L++G A + A+ F + + LF + G + G AG +
Sbjct: 432 LWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAAGASSLLF 611
Query: 122 VCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMARE 181
V + + RL S A G ++ G +DV + ++S+G A GL++G +
Sbjct: 612 VYSLDYARTRLANDSKSAKKGGER---QFNGLVDVYKKTIQSDGIA-GLYRGFSISCIGI 779
Query: 182 VPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQVD 241
+ FG Y++ K ++ G L+ G GA YP D V+ + +
Sbjct: 780 IVYRGLYFGMYDSLKPVVLVGDMQDSFFASFLLGWGITIGAGL--ASYPIDTVRRRMMMT 953
Query: 242 DYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYE 290
+ K+ S+DAF+ I+A EG K L+KG G + R+V A A Y+
Sbjct: 954 SGEAVKYKSSLDAFKVIIAKEGTKSLFKGAGANILRAV-AGAGVLAGYD 1097
>TC7843 weakly similar to GB|AAF03412.1|6090963|AF188712 mitochondrial
dicarboxylate carrier {Mus musculus;} , partial (22%)
Length = 1594
Score = 70.9 bits (172), Expect = 3e-13
Identities = 81/323 (25%), Positives = 130/323 (40%), Gaps = 43/323 (13%)
Frame = +3
Query: 14 GGAAQLIVG---HPFDTIKVKLQSQ---------------------------------PT 37
GG A +I G HP D IKV++Q Q P
Sbjct: 270 GGIASIIAGCSTHPLDLIKVRMQLQGEANAAPKPVQQLRPALAFQNGSTIHVAAHPHSPP 449
Query: 38 PIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGMGAPLATVAALNAVLFTVRGQMEALFRS- 96
P P PR G I + + EG R L+ G+ ATV T G + L +
Sbjct: 450 PPPMPQPRV-GPISVGVRLVQQEGLRALFSGVS---ATVLRQMLYSTTRMGLYDILKQKW 617
Query: 97 HPGASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDV 156
G ++ + +++ G AG + + P ++ R+QA L AA Y +D
Sbjct: 618 SNGGNMPLSRKIEAGLIAGGIGAAVGNPADVAMVRMQADGRL----PAAQRRNYSSVVDA 785
Query: 157 ARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGL---GRGSL 213
+ + E G L++G T+ R + A+ +Y+ K+++ GL G G+
Sbjct: 786 ITRMAKQE-GVTSLWRGSSLTVNRAMLVTASQLASYDQFKEMIL----EKGLMRDGLGTH 950
Query: 214 IVAGGLAGASFWFFVYPTDVVKSVI---QVDDYKYPKFSGSIDAFRRILASEGIKGLYKG 270
+ A AG P DV+K+ + +V+ P ++G++D + + +EG LYKG
Sbjct: 951 VTASFAAGFVAAVASNPVDVIKTRVMNMKVEPGAEPPYAGALDCALKTVRAEGPMALYKG 1130
Query: 271 FGPAMFRSVPANAACFLAYEMTR 293
F P + R P F+ E R
Sbjct: 1131FIPTISRQGPFTVVLFVTLEQVR 1199
Score = 61.6 bits (148), Expect = 2e-10
Identities = 44/195 (22%), Positives = 79/195 (39%)
Frame = +3
Query: 4 VAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPR 63
+++ + AG G VG+P D V++Q+ Q YS +DA+ + EG
Sbjct: 639 LSRKIEAGLIAGGIGAAVGNPADVAMVRMQADGRLPAAQRRNYSSVVDAITRMAKQEGVT 818
Query: 64 GLYKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFLVC 123
L++G + + A Q + + + V AG +
Sbjct: 819 SLWRGSSLTVNRAMLVTASQLASYDQFKEMILEKGLMRDGLGTHVTASFAAGFVAAVASN 998
Query: 124 PTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVP 183
P ++IK R+ G+ Y G +D A +R+E G L+KG +PT++R+ P
Sbjct: 999 PVDVIKTRVMNMKVEPGA-----EPPYAGALDCALKTVRAE-GPMALYKGFIPTISRQGP 1160
Query: 184 GNAAMFGAYEASKQL 198
+F E +++
Sbjct: 1161FTVVLFVTLEQVRKM 1205
Score = 54.7 bits (130), Expect = 2e-08
Identities = 28/90 (31%), Positives = 44/90 (48%)
Frame = +3
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYK 67
+TA G + +P D IK ++ + PG P Y+GA+D +T+ AEGP LYK
Sbjct: 951 VTASFAAGFVAAVASNPVDVIKTRVMNMKVE-PGAEPPYAGALDCALKTVRAEGPMALYK 1127
Query: 68 GMGAPLATVAALNAVLFTVRGQMEALFRSH 97
G ++ VLF Q+ +F+ +
Sbjct: 1128 GFIPTISRQGPFTVVLFVTLEQVRKMFKDY 1217
Score = 47.0 bits (110), Expect = 4e-06
Identities = 37/149 (24%), Positives = 66/149 (43%), Gaps = 4/149 (2%)
Frame = +3
Query: 152 GPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRG 211
GP+ V +++ EG R LF G+ T+ R++ + G Y+ KQ + G +
Sbjct: 477 GPISVGVRLVQQEG-LRALFSGVSATVLRQMLYSTTRMGLYDILKQKWSNG---GNMPLS 644
Query: 212 SLIVAGGLAGASFWFFVYPTDVVKSVIQVDDY----KYPKFSGSIDAFRRILASEGIKGL 267
I AG +AG P DV +Q D + +S +DA R+ EG+ L
Sbjct: 645 RKIEAGLIAGGIGAAVGNPADVAMVRMQADGRLPAAQRRNYSSVVDAITRMAKQEGVTSL 824
Query: 268 YKGFGPAMFRSVPANAACFLAYEMTRSAL 296
++G + R++ A+ +Y+ + +
Sbjct: 825 WRGSSLTVNRAMLVTASQLASYDQFKEMI 911
>TC9482 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier protein-like
(At5g48970), partial (42%)
Length = 529
Score = 68.6 bits (166), Expect = 1e-12
Identities = 47/145 (32%), Positives = 69/145 (47%), Gaps = 18/145 (12%)
Frame = +3
Query: 166 GARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGG-----PDTSGLGRGS---LIVAG 217
G +G++ GL PT+ +P FG Y+ K+ +TS S L + G
Sbjct: 9 GFQGMYAGLSPTLVEIIPYAGLQFGTYDTFKRWAMAWNHYRYSNTSAENSPSSFQLFLCG 188
Query: 218 GLAGASFWFFVYPTDVVKSVIQVDDY-KYPKF---------SGSIDAFRRILASEGIKGL 267
AG +P DVVK Q++ ++P++ S DA +RIL EG GL
Sbjct: 189 LAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYSSMFDAIQRILRLEGWAGL 368
Query: 268 YKGFGPAMFRSVPANAACFLAYEMT 292
YKG P+ ++ PA A F+AYE+T
Sbjct: 369 YKGIIPSTVKAAPAGAVTFVAYELT 443
Score = 48.9 bits (115), Expect = 1e-06
Identities = 29/96 (30%), Positives = 42/96 (43%), Gaps = 2/96 (2%)
Frame = +3
Query: 106 QQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGS--GTAAVAVKYGGPMDVARHVLRS 163
Q CG AG + P +++K R Q + G Y D + +LR
Sbjct: 171 QLFLCGLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYSSMFDAIQRILRL 350
Query: 164 EGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLL 199
EG A GL+KG++P+ + P A F AYE + L
Sbjct: 351 EGWA-GLYKGIIPSTVKAAPAGAVTFVAYELTSDWL 455
Score = 37.7 bits (86), Expect = 0.002
Identities = 27/80 (33%), Positives = 35/80 (43%), Gaps = 6/80 (7%)
Frame = +3
Query: 11 GTFGGAAQLIVGHPFDTIKVKLQS---QPTPIPG---QLPRYSGAIDAVKQTLAAEGPRG 64
G G +V HP D +K + Q Q P G + YS DA+++ L EG G
Sbjct: 186 GLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYSSMFDAIQRILRLEGWAG 365
Query: 65 LYKGMGAPLATVAALNAVLF 84
LYKG+ A AV F
Sbjct: 366 LYKGIIPSTVKAAPAGAVTF 425
>BP047276
Length = 516
Score = 62.4 bits (150), Expect = 9e-11
Identities = 49/157 (31%), Positives = 73/157 (46%), Gaps = 2/157 (1%)
Frame = +3
Query: 26 DTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGMGAPLATVAALN-AVLF 84
D IK +LQ + Y G + EG R L+KG+ P AT A+
Sbjct: 84 DVIKTRLQLDRSG------NYKGIVHCGSTISRTEGVRALWKGL-TPFATHLTFKYALRM 242
Query: 85 TVRGQMEALFRSHPGASLTVHQQVFCGAGAG-LAVSFLVCPTELIKCRLQAQSTLAGSGT 143
+++F+ L+ H ++ G GAG L +V P E++K +LQ Q G
Sbjct: 243 GSNAVFQSMFKDSETGKLSSHGRLLSGFGAGVLEAIVIVTPFEVVKIKLQQQR-----GL 407
Query: 144 AAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAR 180
+ +KY GP+ AR +L E G RGL+ G+ PT+ R
Sbjct: 408 SPELLKYKGPVHCARTILHEE-GIRGLWAGVSPTIMR 515
Score = 37.4 bits (85), Expect = 0.003
Identities = 18/51 (35%), Positives = 30/51 (58%)
Frame = +3
Query: 19 LIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGM 69
+++ PF+ +K+KLQ Q P +L +Y G + + L EG RGL+ G+
Sbjct: 348 IVIVTPFEVVKIKLQQQRGLSP-ELLKYKGPVHCARTILHEEGIRGLWAGV 497
Score = 28.1 bits (61), Expect = 1.9
Identities = 14/48 (29%), Positives = 26/48 (54%)
Frame = +3
Query: 226 FFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGP 273
F + DV+K+ +Q+D + + G + I +EG++ L+KG P
Sbjct: 66 FLLATHDVIKTRLQLD--RSGNYKGIVHCGSTISRTEGVRALWKGLTP 203
>TC7883 homologue to UP|ADT1_GOSHI (O22342) ADP,ATP carrier protein 1,
mitochondrial precursor (ADP/ATP translocase 1) (Adenine
nucleotide translocator 1) (ANT 1), partial (54%)
Length = 856
Score = 62.0 bits (149), Expect = 1e-10
Identities = 51/171 (29%), Positives = 76/171 (43%), Gaps = 8/171 (4%)
Frame = +2
Query: 109 FCGAGAGLAVS-FLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGA 167
F G AVS P E +K +Q Q + SG ++ Y G D ++ EG
Sbjct: 284 FLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKSGR--LSEPYKGISDCFGRTMKDEG-V 454
Query: 168 RGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGR--GSLIVAGGLAGASFW 225
L++G + R P A F + K+L D G + + +GG AGAS
Sbjct: 455 IALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAAGASSL 634
Query: 226 FFVYPTDVVKSVIQVDDYKYPK-----FSGSIDAFRRILASEGIKGLYKGF 271
FVY D ++ + D K F+G ID +++ + S+GI GLY+GF
Sbjct: 635 LFVYSLDYARTRLANDAKAAKKGGERQFNGLIDVYKKTIKSDGIAGLYRGF 787
>BU494115
Length = 466
Score = 40.0 bits (92), Expect(2) = 1e-09
Identities = 22/63 (34%), Positives = 33/63 (51%)
Frame = +2
Query: 131 RLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFG 190
R+Q Q T ++ +Y P+D A +++EG G+F+G T+ RE GNA F
Sbjct: 11 RVQIQGT---DSLVPMSSRYSSPLDCALKTVKNEG-VTGIFRGGCATLLRESMGNATFFS 178
Query: 191 AYE 193
YE
Sbjct: 179 VYE 187
Score = 38.1 bits (87), Expect(2) = 1e-09
Identities = 25/66 (37%), Positives = 35/66 (52%), Gaps = 1/66 (1%)
Frame = +3
Query: 204 DTSGL-GRGSLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASE 262
D SGL G IV GG++G +FW V P DV K++IQ + K + + + I
Sbjct: 234 DYSGLIDIGIGIVTGGVSGIAFWLAVLPLDVAKTLIQTNPDKNSQRNPFV-ILSSIYKRA 410
Query: 263 GIKGLY 268
G+KG Y
Sbjct: 411 GLKGCY 428
>TC14563 similar to UP|Q9SSP4 (Q9SSP4) F3N23.2 protein (At1g72820/F3N23_2),
partial (55%)
Length = 1025
Score = 58.2 bits (139), Expect = 2e-09
Identities = 55/194 (28%), Positives = 84/194 (42%), Gaps = 13/194 (6%)
Frame = +2
Query: 109 FCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGAR 168
F GA VS + P ++K R Q VA + + A ++R EG R
Sbjct: 479 FLGAALFSGVSATLYPAVVLKTRQQ------------VAQTHVSCIKTAFTLIRGEG-FR 619
Query: 169 GLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIV----AGGLAGASF 224
L++G ++ +P A A E +K + G + A GL+ A
Sbjct: 620 ALYRGFGTSLMGTIPARALYMAALEVTKSNVGTATVRFGFAEPTAAAIASAAAGLSAAMA 799
Query: 225 WFFVY-PTDVVKSVIQVD---DYKYPKFSG-----SIDAFRRILASEGIKGLYKGFGPAM 275
V+ P DVV + V + PK S IDAFR+IL ++G +GLY+GFG ++
Sbjct: 800 AQLVWTPVDVVSQRLMVQGCCNSSNPKGSAVRYINGIDAFRKILNTDGPRGLYRGFGISI 979
Query: 276 FRSVPANAACFLAY 289
P+NA + +Y
Sbjct: 980 LTYAPSNAVWWASY 1021
Score = 46.6 bits (109), Expect = 5e-06
Identities = 28/77 (36%), Positives = 37/77 (47%), Gaps = 4/77 (5%)
Frame = +2
Query: 10 AGTFGGAAQLIVGHPFDTIKVKLQSQ----PTPIPGQLPRYSGAIDAVKQTLAAEGPRGL 65
AG A +V P D + +L Q + G RY IDA ++ L +GPRGL
Sbjct: 776 AGLSAAMAAQLVWTPVDVVSQRLMVQGCCNSSNPKGSAVRYINGIDAFRKILNTDGPRGL 955
Query: 66 YKGMGAPLATVAALNAV 82
Y+G G + T A NAV
Sbjct: 956 YRGFGISILTYAPSNAV 1006
Score = 38.5 bits (88), Expect = 0.001
Identities = 22/70 (31%), Positives = 36/70 (51%)
Frame = +2
Query: 228 VYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFL 287
+YP V+K+ QV I ++ EG + LY+GFG ++ ++PA A
Sbjct: 518 LYPAVVLKTRQQVAQTHV----SCIKTAFTLIRGEGFRALYRGFGTSLMGTIPARALYMA 685
Query: 288 AYEMTRSALG 297
A E+T+S +G
Sbjct: 686 ALEVTKSNVG 715
>TC8324 UP|Q8VWY1 (Q8VWY1) Mitochondrial phosphate transporter, complete
Length = 1605
Score = 57.8 bits (138), Expect = 2e-09
Identities = 58/196 (29%), Positives = 84/196 (42%), Gaps = 5/196 (2%)
Frame = +3
Query: 100 ASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARH 159
A+ TV + CG V P +L+KC +Q T S ++ +G
Sbjct: 330 AACTVGGILSCGL-----THMTVTPLDLVKCNMQIDPTKYKS----ISSGFG-------- 458
Query: 160 VLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLA--GGPDTSGLGRGSLIVAG 217
VL E G +G F+G VPT+ A FG YE K+ + GP+ + + +LI
Sbjct: 459 VLLKEQGVKGFFRGWVPTLLGYSAQGACKFGFYEFFKKYYSDIAGPEYATKYK-TLIYLA 635
Query: 218 GLAGASFWFFV--YPTDVVKSVIQVDDYKYPKFS-GSIDAFRRILASEGIKGLYKGFGPA 274
G A A V P + VK +Q P F+ G D + + +EG GLYKG P
Sbjct: 636 GSASAEVIADVALCPFEAVKVRVQTQ----PGFARGLSDGLPKFVKAEGTLGLYKGLVPL 803
Query: 275 MFRSVPANAACFLAYE 290
R +P F ++E
Sbjct: 804 WGRQIPYTMMKFASFE 851
Score = 48.5 bits (114), Expect = 1e-06
Identities = 46/198 (23%), Positives = 73/198 (36%), Gaps = 8/198 (4%)
Frame = +3
Query: 10 AGTFGGAAQLIVGH----PFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGL 65
A T GG + H P D +K +Q PT +Y L +G +G
Sbjct: 333 ACTVGGILSCGLTHMTVTPLDLVKCNMQIDPT-------KYKSISSGFGVLLKEQGVKGF 491
Query: 66 YKGMGAPLATVAALNAVLFTVRGQMEALFRSHPGASLTVHQQVFC----GAGAGLAVSFL 121
++G L +A A F + + G + A A +
Sbjct: 492 FRGWVPTLLGYSAQGACKFGFYEFFKKYYSDIAGPEYATKYKTLIYLAGSASAEVIADVA 671
Query: 122 VCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMARE 181
+CP E +K R+Q Q A G D +++EG GL+KGLVP R+
Sbjct: 672 LCPFEAVKVRVQTQPGFAR-----------GLSDGLPKFVKAEG-TLGLYKGLVPLWGRQ 815
Query: 182 VPGNAAMFGAYEASKQLL 199
+P F ++E +++
Sbjct: 816 IPYTMMKFASFETIVEMI 869
>TC18745 weakly similar to UP|ADT2_HUMAN (P05141) ADP,ATP carrier protein,
fibroblast isoform (ADP/ATP translocase 2) (Adenine
nucleotide translocator 2) (ANT 2), partial (9%)
Length = 834
Score = 57.0 bits (136), Expect = 4e-09
Identities = 43/133 (32%), Positives = 59/133 (44%), Gaps = 5/133 (3%)
Frame = +3
Query: 163 SEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAG--GPDTSGLGRGSLIV---AG 217
+E G R KG + T+ +P +A F +YE K L G + G +L V G
Sbjct: 435 NEEGFRAFGKGNMVTIVHRLPYSAVSFYSYERYKNLFHSLMGENYRGSSSANLFVHFMGG 614
Query: 218 GLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFR 277
GLAG + YP D+V++ + + G AF I EG GLYKG G +
Sbjct: 615 GLAGVTSASATYPLDLVRTRLAAQRSTI-YYRGISHAFSTICRDEGFLGLYKGLGATLLG 791
Query: 278 SVPANAACFLAYE 290
P+ A F YE
Sbjct: 792 VGPSIAISFSVYE 830
Score = 40.4 bits (93), Expect = 4e-04
Identities = 52/196 (26%), Positives = 72/196 (36%), Gaps = 9/196 (4%)
Frame = +3
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYK 67
L AG GA P + + Q Q R + + EG R K
Sbjct: 285 LLAGGVAGAFSKTCTAPLARLTILFQVQGMHSDFSALRKPSIWQEASRIINEEGFRAFGK 464
Query: 68 GMGAPLATVAALNAVLFTVRGQMEALFRSHPG--------ASLTVHQQVFCGAG-AGLAV 118
G + +AV F + + LF S G A+L VH F G G AG+
Sbjct: 465 GNMVTIVHRLPYSAVSFYSYERYKNLFHSLMGENYRGSSSANLFVH---FMGGGLAGVTS 635
Query: 119 SFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTM 178
+ P +L++ RL AQ + + Y G + R EG GL+KGL T+
Sbjct: 636 ASATYPLDLVRTRLAAQRS---------TIYYRGISHAFSTICRDEGFL-GLYKGLGATL 785
Query: 179 AREVPGNAAMFGAYEA 194
P A F YE+
Sbjct: 786 LGVGPSIAISFSVYES 833
>AV408895
Length = 429
Score = 54.7 bits (130), Expect = 2e-08
Identities = 41/137 (29%), Positives = 60/137 (42%), Gaps = 6/137 (4%)
Frame = +3
Query: 163 SEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLA------GGPDTSGLGRGSLIVA 216
+E G R +KG + T+A +P ++ F +YE K+ L D V
Sbjct: 21 NEEGVRAFWKGNLVTIAHRLPYSSVNFYSYEHYKKWLRMVSGVQNHRDNVSADVFIHFVG 200
Query: 217 GGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMF 276
GG+AG + YP D+V++ + + + G A + I EGI GLYKG G +
Sbjct: 201 GGMAGITAATSTYPLDLVRTRLAAQT-NFTYYRGIWHALQTISKEEGILGLYKGLGTTLL 377
Query: 277 RSVPANAACFLAYEMTR 293
P A F YE R
Sbjct: 378 TVGPNIAISFSVYESLR 428
Score = 37.4 bits (85), Expect = 0.003
Identities = 25/84 (29%), Positives = 35/84 (40%)
Frame = +3
Query: 3 DVAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGP 62
DV G G +P D ++ +L +Q Y G A++ EG
Sbjct: 177 DVFIHFVGGGMAGITAATSTYPLDLVRTRLAAQTN-----FTYYRGIWHALQTISKEEGI 341
Query: 63 RGLYKGMGAPLATVAALNAVLFTV 86
GLYKG+G L TV A+ F+V
Sbjct: 342 LGLYKGLGTTLLTVGPNIAISFSV 413
Score = 35.8 bits (81), Expect = 0.009
Identities = 29/101 (28%), Positives = 43/101 (41%), Gaps = 1/101 (0%)
Frame = +3
Query: 95 RSHPGASLTVHQQVFCGAG-AGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGP 153
R + A + +H F G G AG+ + P +L++ RL AQ+ Y
Sbjct: 159 RDNVSADVFIH---FVGGGMAGITAATSTYPLDLVRTRLAAQTNFT----------YYRG 299
Query: 154 MDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEA 194
+ A + E G GL+KGL T+ P A F YE+
Sbjct: 300 IWHALQTISKEEGILGLYKGLGTTLLTVGPNIAISFSVYES 422
>CB828090
Length = 427
Score = 53.1 bits (126), Expect = 5e-08
Identities = 29/83 (34%), Positives = 44/83 (52%), Gaps = 10/83 (12%)
Frame = +2
Query: 220 AGASFWFFVYPTDVVKSVIQVDDY-KYPKFSGSI---------DAFRRILASEGIKGLYK 269
AG +P DVVK Q++ ++P++ + DA +RI+ EG GLYK
Sbjct: 5 AGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAYKNMFDAMKRIIQMEGWAGLYK 184
Query: 270 GFGPAMFRSVPANAACFLAYEMT 292
G P+ ++ PA A F+AYE+T
Sbjct: 185 GLFPSTVKAAPAGAVTFVAYELT 253
Score = 39.3 bits (90), Expect = 8e-04
Identities = 25/88 (28%), Positives = 38/88 (42%), Gaps = 2/88 (2%)
Frame = +2
Query: 114 AGLAVSFLVCPTELIKCRLQAQSTLAGS--GTAAVAVKYGGPMDVARHVLRSEGGARGLF 171
AG + P +++K R Q + G Y D + +++ EG A GL+
Sbjct: 5 AGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAYKNMFDAMKRIIQMEGWA-GLY 181
Query: 172 KGLVPTMAREVPGNAAMFGAYEASKQLL 199
KGL P+ + P A F AYE + L
Sbjct: 182 KGLFPSTVKAAPAGAVTFVAYELTSDWL 265
Score = 35.8 bits (81), Expect = 0.009
Identities = 24/79 (30%), Positives = 34/79 (42%), Gaps = 9/79 (11%)
Frame = +2
Query: 15 GAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAI---------DAVKQTLAAEGPRGL 65
G +V HP D +K + Q + + PRY + DA+K+ + EG GL
Sbjct: 8 GTCAKLVCHPLDVVKKRFQIEGLQ---RHPRYGARVEHRAYKNMFDAMKRIIQMEGWAGL 178
Query: 66 YKGMGAPLATVAALNAVLF 84
YKG+ A AV F
Sbjct: 179YKGLFPSTVKAAPAGAVTF 235
>TC17966 similar to UP|Q8L9F8 (Q8L9F8) Mitochondrial carrier-like protein,
partial (53%)
Length = 917
Score = 52.8 bits (125), Expect = 7e-08
Identities = 51/210 (24%), Positives = 78/210 (36%), Gaps = 21/210 (10%)
Frame = +2
Query: 10 AGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQL------PRYSGAIDAVKQTLAAEGPR 63
AG A +V P D + +L Q T + Y DA ++ L A+GP+
Sbjct: 80 AGVASAMAAQLVWTPIDVVSQRLMVQGTGGSKTMLANLNSESYRNGFDAFRKILCADGPK 259
Query: 64 GLYKGMGAPLATVAALNAVLFT---------------VRGQMEALFRSHPGASLTVHQQV 108
G Y+G G L T A NAV +T GQ + + P + V Q
Sbjct: 260 GFYRGFGISLLTYAPSNAVWWTSYSMVHRFIWGSFGPYMGQRDNVCGFRPDSKSIVAVQG 439
Query: 109 FCGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGAR 168
A + + P + I+ RLQ T + + R++++ EGG
Sbjct: 440 LSAVVASGVSAIVTMPLDTIQTRLQVLDTEENGRRRPLT-----SVQTLRNLVK-EGGLL 601
Query: 169 GLFKGLVPTMAREVPGNAAMFGAYEASKQL 198
++GL P A M YE K++
Sbjct: 602 ACYRGLGPRWASMSMSATTMITTYEFVKRM 691
Score = 45.8 bits (107), Expect = 9e-06
Identities = 27/87 (31%), Positives = 40/87 (45%), Gaps = 10/87 (11%)
Frame = +2
Query: 215 VAGGLAGASFWFFVYPTDVVKSVIQVD----------DYKYPKFSGSIDAFRRILASEGI 264
VA +A W P DVV + V + + DAFR+IL ++G
Sbjct: 86 VASAMAAQLVWT---PIDVVSQRLMVQGTGGSKTMLANLNSESYRNGFDAFRKILCADGP 256
Query: 265 KGLYKGFGPAMFRSVPANAACFLAYEM 291
KG Y+GFG ++ P+NA + +Y M
Sbjct: 257 KGFYRGFGISLLTYAPSNAVWWTSYSM 337
Score = 40.0 bits (92), Expect = 5e-04
Identities = 42/207 (20%), Positives = 79/207 (37%), Gaps = 24/207 (11%)
Frame = +2
Query: 111 GAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVK---YGGPMDVARHVLRSEGGA 167
G + +A + P +++ RL Q T GS T + Y D R +L ++G
Sbjct: 83 GVASAMAAQLVWTPIDVVSQRLMVQGT-GGSKTMLANLNSESYRNGFDAFRKILCADG-P 256
Query: 168 RGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGG---------------PDTSGLGRG- 211
+G ++G ++ P NA + +Y + + G PD+ +
Sbjct: 257 KGFYRGFGISLLTYAPSNAVWWTSYSMVHRFIWGSFGPYMGQRDNVCGFRPDSKSIVAVQ 436
Query: 212 --SLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYKYP---KFSGSIDAFRRILASEGIKG 266
S +VA G++ P D +++ +QV D + + S+ R ++ G+
Sbjct: 437 GLSAVVASGVSA----IVTMPLDTIQTRLQVLDTEENGRRRPLTSVQTLRNLVKEGGLLA 604
Query: 267 LYKGFGPAMFRSVPANAACFLAYEMTR 293
Y+G GP + YE +
Sbjct: 605 CYRGLGPRWASMSMSATTMITTYEFVK 685
>TC8441 homologue to UP|ADT2_SOLTU (P27081) ADP,ATP carrier protein,
mitochondrial precursor (ADP/ATP translocase) (Adenine
nucleotide translocator) (ANT) (Fragment), partial (48%)
Length = 884
Score = 52.8 bits (125), Expect = 7e-08
Identities = 42/130 (32%), Positives = 63/130 (48%), Gaps = 5/130 (3%)
Frame = +1
Query: 166 GARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLAGGP--DT--SGLGRGSLIVAG-GLA 220
G GL++G + + FG Y++ K +L G D+ + G G LI G GLA
Sbjct: 157 GVAGLYRGFNISCVGIIVYRGLYFGMYDSLKPVLLTGSLQDSFFASFGLGWLITNGAGLA 336
Query: 221 GASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVP 280
YP D V+ + + + K+ S+DAF +IL +EG K L+KG G + R+V
Sbjct: 337 S-------YPIDTVRRRMMMTSGEAVKYKSSMDAFTQILKNEGPKSLFKGAGANILRAV- 492
Query: 281 ANAACFLAYE 290
A A Y+
Sbjct: 493 AGAGVLAGYD 522
Score = 39.3 bits (90), Expect = 8e-04
Identities = 20/64 (31%), Positives = 33/64 (51%)
Frame = +1
Query: 20 IVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYKGMGAPLATVAAL 79
+ +P DT++ ++ G+ +Y ++DA Q L EGP+ L+KG GA + A
Sbjct: 331 LASYPIDTVRRRMMMTS----GEAVKYKSSMDAFTQILKNEGPKSLFKGAGANILRAVAG 498
Query: 80 NAVL 83
VL
Sbjct: 499 AGVL 510
>TC9406 similar to UP|Q9M058 (Q9M058) Ca-dependent solute carrier-like
protein, partial (30%)
Length = 676
Score = 52.4 bits (124), Expect = 9e-08
Identities = 34/123 (27%), Positives = 59/123 (47%)
Frame = +3
Query: 81 AVLFTVRGQMEALFRSHPGASLTVHQQVFCGAGAGLAVSFLVCPTELIKCRLQAQSTLAG 140
A+ F+V + +L+ S + + C + +G+A S ++ P +L++ R+Q +
Sbjct: 9 AISFSVYESLRSLWISRRPDDSSAVVSLACASLSGVASSTVIFPLDLVRRRMQLE----- 173
Query: 141 SGTAAVAVKYGGPMDVARHVLRSEGGARGLFKGLVPTMAREVPGNAAMFGAYEASKQLLA 200
G A Y M A + G RGL++G++P + VPG F YE K+LL+
Sbjct: 174 -GVGGRACVYNTGMCGAFQCIIKNEGVRGLYRGILPEYYKVVPGVGIAFMTYETLKRLLS 350
Query: 201 GGP 203
P
Sbjct: 351 STP 359
Score = 47.0 bits (110), Expect = 4e-06
Identities = 31/115 (26%), Positives = 56/115 (47%), Gaps = 4/115 (3%)
Frame = +3
Query: 186 AAMFGAYEASKQL-LAGGPDTSGLGRGSLIVAGGLAGASFWFFVYPTDVVKSVIQVDDYK 244
A F YE+ + L ++ PD S + L+G + ++P D+V+ +Q++
Sbjct: 9 AISFSVYESLRSLWISRRPDDSSAVVS--LACASLSGVASSTVIFPLDLVRRRMQLEGVG 182
Query: 245 YPKF---SGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACFLAYEMTRSAL 296
+G AF+ I+ +EG++GLY+G P ++ VP F+ YE + L
Sbjct: 183 GRACVYNTGMCGAFQCIIKNEGVRGLYRGILPEYYKVVPGVGIAFMTYETLKRLL 347
Score = 27.7 bits (60), Expect = 2.4
Identities = 23/92 (25%), Positives = 39/92 (42%), Gaps = 1/92 (1%)
Frame = +3
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYS-GAIDAVKQTLAAEGPRGLY 66
L + G A V P D ++ ++Q + + G+ Y+ G A + + EG RGLY
Sbjct: 90 LACASLSGVASSTVIFPLDLVRRRMQLEG--VGGRACVYNTGMCGAFQCIIKNEGVRGLY 263
Query: 67 KGMGAPLATVAALNAVLFTVRGQMEALFRSHP 98
+G+ V + F ++ L S P
Sbjct: 264 RGILPEYYKVVPGVGIAFMTYETLKRLLSSTP 359
>TC7986 similar to UP|Q8SF02 (Q8SF02) Dicarboxylate/tricarboxylate carrier,
partial (96%)
Length = 1532
Score = 52.0 bits (123), Expect = 1e-07
Identities = 43/186 (23%), Positives = 79/186 (42%), Gaps = 4/186 (2%)
Frame = +3
Query: 111 GAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARGL 170
G +G+ + ++ P ++IK R+Q G G+A + V ++L+++G
Sbjct: 252 GGASGMLATCVIQPIDMIKVRIQL-----GQGSA---------VQVTSNMLKNDG-VGAF 386
Query: 171 FKGLVPTMAREVPGNAAMFGAYEASKQLLAGGPDTSGLGRGSLIVAGGLAGASFWFFVYP 230
+KGL + R+ A G + D L + G AGA P
Sbjct: 387 YKGLSAGLLRQATYTTARLGTFRILTSKAIEANDGKPLPLYQKALCGLTAGAIGATVGSP 566
Query: 231 TDVVKSVIQVDD----YKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFRSVPANAACF 286
D+ +Q D ++ ++ + A RI+A EG+ L+KG GP + R++ N
Sbjct: 567 ADLALIRMQADATLPLHQRRHYTNAFHALYRIVADEGVLSLWKGAGPTVVRAMALNMGML 746
Query: 287 LAYEMT 292
+Y+ +
Sbjct: 747 ASYDQS 764
Score = 47.8 bits (112), Expect = 2e-06
Identities = 44/153 (28%), Positives = 65/153 (41%), Gaps = 7/153 (4%)
Frame = +3
Query: 8 LTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEGPRGLYK 67
LTAG G VG P D +++Q+ T Q Y+ A A+ + +A EG L+K
Sbjct: 528 LTAGAIGAT----VGSPADLALIRMQADATLPLHQRRHYTNAFHALYRIVADEGVLSLWK 695
Query: 68 GMGAPLATVAALNAVLFTVRGQMEALFRSHPG---ASLTVHQQVFCG-AGAGLAVSFLVC 123
G G + ALN + Q F+ G AS V G A ++ F
Sbjct: 696 GAGPTVVRAMALNMGMLASYDQSVEFFKDSVGLGEASTVVAASSVSGFFAAACSLPFDYV 875
Query: 124 PTELIKCRLQAQSTLAGSGT---AAVAVKYGGP 153
T++ K + A+ +G+ AA +K GGP
Sbjct: 876 KTQIQKMQPDAEGKYPYTGSLDCAAKTLKQGGP 974
>TC19374 similar to UP|Q8LNZ1 (Q8LNZ1) Mitochondrial uncoupling protein,
partial (49%)
Length = 556
Score = 51.6 bits (122), Expect = 2e-07
Identities = 38/140 (27%), Positives = 60/140 (42%), Gaps = 5/140 (3%)
Frame = +3
Query: 5 AKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQ---LPRYSGAIDAVKQTLAAEG 61
AK + F + P DT KV+LQ Q I G LP+Y G + + EG
Sbjct: 120 AKTFASSAFSACFAEVCTIPLDTAKVRLQLQKQGIAGDVASLPKYKGMLGTIATIAREEG 299
Query: 62 PRGLYKGMGAPLATVAALNAVLFTVRGQMEALF--RSHPGASLTVHQQVFCGAGAGLAVS 119
L+KG+ L + + ++AL+ H G + + +++ G
Sbjct: 300 ASALWKGIVPGLHRQCLYGGLRIGLYEPVKALYVGSDHVG-DVPLSKKILAAFTTGAVAI 476
Query: 120 FLVCPTELIKCRLQAQSTLA 139
+ PT+L+K RLQA+ LA
Sbjct: 477 TVANPTDLVKVRLQAEGKLA 536
Score = 27.7 bits (60), Expect = 2.4
Identities = 14/43 (32%), Positives = 18/43 (41%)
Frame = +3
Query: 235 KSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPAMFR 277
K I D PK+ G + I EG L+KG P + R
Sbjct: 213 KQGIAGDVASLPKYKGMLGTIATIAREEGASALWKGIVPGLHR 341
>TC10218 similar to UP|Q9SJ70 (Q9SJ70) Probable calcium-binding
mitochondrial carrier At2g35800, partial (28%)
Length = 709
Score = 51.6 bits (122), Expect = 2e-07
Identities = 66/250 (26%), Positives = 92/250 (36%), Gaps = 15/250 (6%)
Frame = +1
Query: 2 GDVAKDLTAGTFGGAAQLIVGHPFDTIKVKLQSQPTPIPGQLPRYSGAIDAVKQTLAAEG 61
G V + AG A + HP D+IK ++Q+ P + + L G
Sbjct: 79 GSVLRSALAGGLACALSCALLHPVDSIKTRVQASTMSFPEIISK-----------LPQIG 225
Query: 62 PRGLYKGMGAPLATVAALNAVL--FTVRGQMEALFRSHPGASLTVHQQV----------F 109
RGLY+G ++ A+L F+ G +F + + V + F
Sbjct: 226 VRGLYRG---------SIPAILGQFSSHGLRTGIFEASKLVMINVAPTLPELQVQSVASF 378
Query: 110 CGAGAGLAVSFLVCPTELIKCRLQAQSTLAGSGTAAVAVKYGGPMDVARHVLRSEGGARG 169
C G AV P E++K RLQA G A V + G +G
Sbjct: 379 CSTFLGTAVRI---PCEVLKQRLQA-GIFNNVGEALVGTW-------------EQDGVKG 507
Query: 170 LFKGLVPTMAREVPGNAAMFGAYEASK---QLLAGGPDTSGLGRGSLIVAGGLAGASFWF 226
F+G T+ REVP A G Y SK Q L G L I G L+G
Sbjct: 508 FFRGTGATLCREVPFYVAGMGLYAESKKGVQKLLG----RELEAWETIAVGALSGGLAAV 675
Query: 227 FVYPTDVVKS 236
P DV+K+
Sbjct: 676 VTTPFDVMKT 705
Score = 34.3 bits (77), Expect = 0.026
Identities = 19/61 (31%), Positives = 31/61 (50%)
Frame = +1
Query: 215 VAGGLAGASFWFFVYPTDVVKSVIQVDDYKYPKFSGSIDAFRRILASEGIKGLYKGFGPA 274
+AGGLA A ++P D +K+ +Q +P+ L G++GLY+G PA
Sbjct: 100 LAGGLACALSCALLHPVDSIKTRVQASTMSFPEIISK-------LPQIGVRGLYRGSIPA 258
Query: 275 M 275
+
Sbjct: 259 I 261
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,205,135
Number of Sequences: 28460
Number of extensions: 49153
Number of successful extensions: 363
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 280
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 336
length of query: 297
length of database: 4,897,600
effective HSP length: 90
effective length of query: 207
effective length of database: 2,336,200
effective search space: 483593400
effective search space used: 483593400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0381.7


