

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0380.6
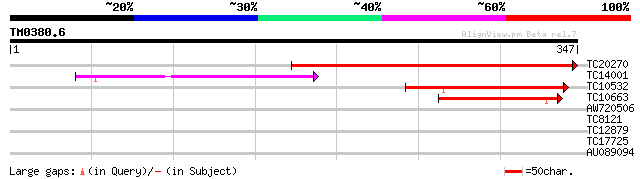
(347 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC20270 similar to UP|O65494 (O65494) UDP-galactose transporter ... 344 1e-95
TC14001 similar to UP|Q8LGE9 (Q8LGE9) CMP-sialic acid transporte... 115 1e-26
TC10532 similar to UP|Q8LGE9 (Q8LGE9) CMP-sialic acid transporte... 91 3e-19
TC10663 similar to UP|Q8VYH1 (Q8VYH1) AT5g65000/MXK3_23, partial... 45 2e-05
AW720506 33 0.071
TC8121 UP|Q9LKJ5 (Q9LKJ5) Multifunctional transport intrinsic me... 28 2.3
TC12879 27 3.9
TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, part... 27 6.6
AU089094 27 6.6
>TC20270 similar to UP|O65494 (O65494) UDP-galactose transporter - like
protein (UDP-galactose transporter-like protein),
partial (98%)
Length = 768
Score = 344 bits (883), Expect = 1e-95
Identities = 174/175 (99%), Positives = 174/175 (99%)
Frame = +1
Query: 173 QWAAFILLTAGCTTAQLNSNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRN 232
QWAAFILLTAGCTTAQLNSNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRN
Sbjct: 1 QWAAFILLTAGCTTAQLNSNSDHVLQTPFQGWVMAIAMALLSGFAGVYTEAIIKKRPSRN 180
Query: 233 INVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMK 292
INVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMK
Sbjct: 181 INVQNFWLYVFGMGFNAVAILVQDFDAVMNKGFFHGYSFITVLMIFNHALSGIAVSMVMK 360
Query: 293 YADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLHSAGKMQR 347
YADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVA YLHSAGKMQR
Sbjct: 361 YADNIVKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAFYLHSAGKMQR 525
>TC14001 similar to UP|Q8LGE9 (Q8LGE9) CMP-sialic acid transporter-like
protein, partial (43%)
Length = 497
Score = 115 bits (287), Expect = 1e-26
Identities = 61/151 (40%), Positives = 90/151 (59%), Gaps = 2/151 (1%)
Frame = +3
Query: 41 QWKRKSMVTFA--LTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVALGRI 98
+WK+ + A LT+LTSSQ IL S+ GKY+Y T F+ E K A+S + L
Sbjct: 12 KWKKMQWYSVAALLTVLTSSQGILTTLSQTNGKYDYDYATIPFLAEVFKLAVSCLLL--- 182
Query: 99 WNKEGVTDDNRLTTTLDEVIVYPIPAALYLVKNLLQYYIFAYVDAPGYQILKNFNIISTG 158
W + + ++TT V +YPIP+ +YL+ N +Q+ YVD YQI+ N I++TG
Sbjct: 183 WRECKSSPLPKMTTDWKTVALYPIPSVIYLIHNNVQFATLMYVDTSTYQIMGNLKIVTTG 362
Query: 159 VLYRIILKKRLSEIQWAAFILLTAGCTTAQL 189
+L+R+ L +RLS +QW A LL G TT+Q+
Sbjct: 363 ILFRLFLGRRLSNLQWMAVGLLAIGTTTSQV 455
>TC10532 similar to UP|Q8LGE9 (Q8LGE9) CMP-sialic acid transporter-like
protein, partial (34%)
Length = 750
Score = 90.9 bits (224), Expect = 3e-19
Identities = 47/105 (44%), Positives = 68/105 (64%), Gaps = 5/105 (4%)
Frame = +3
Query: 243 FGMGFNAVAILVQDFDAVMNKG-----FFHGYSFITVLMIFNHALSGIAVSMVMKYADNI 297
FG FN ++V DF G F+GY+ T L++ N +G+ VS +MKYADNI
Sbjct: 63 FGTIFNLARLVVDDFRGGFENGPWWQRIFNGYTITTWLVVLNLGSTGLLVSWLMKYADNI 242
Query: 298 VKVYSTSVAMLLTAVVSVFLFGFHLSLAFFLGTIVVSVAIYLHSA 342
VKVYSTS+AMLLT V+S+FLF F +L FLG ++ ++++++ A
Sbjct: 243 VKVYSTSMAMLLTMVLSIFLFDFKPTLQLFLGIVICMMSLHMYFA 377
>TC10663 similar to UP|Q8VYH1 (Q8VYH1) AT5g65000/MXK3_23, partial (27%)
Length = 584
Score = 45.1 bits (105), Expect = 2e-05
Identities = 25/78 (32%), Positives = 47/78 (60%), Gaps = 2/78 (2%)
Frame = +1
Query: 263 KGFFHGYSFITVLMIFNHALSGIAVSMVMKYADNIVKVYSTSVAMLLTAVVSVFLFGFHL 322
+GF +G++ +T++ + +HAL+GI V +V +YA + K + A+L+TA++ G
Sbjct: 10 RGFCYGWTPLTLVPVISHALAGILVGLVTRYAGGVRKGFVLVSALLVTALLPFLFEGKPP 189
Query: 323 SLAFF--LGTIVVSVAIY 338
SL L +V S+++Y
Sbjct: 190 SLYCLAALPLVVSSISVY 243
>AW720506
Length = 572
Score = 33.1 bits (74), Expect = 0.071
Identities = 22/73 (30%), Positives = 36/73 (49%), Gaps = 4/73 (5%)
Frame = +2
Query: 19 SVAVNNTVSDGETKVDSHRTQVQWKRKSMVTFALT----ILTSSQAILIVWSKRAGKYEY 74
S V+ T D H++++ K++ ++ F L IL Q IL+ SK GK+ +
Sbjct: 287 SKLVSPTTKSISRAYDRHKSRITSKQR-VLNFLLVGGDCILVGLQPILVYMSKVDGKFNF 463
Query: 75 SVTTANFMVETLK 87
S + NF+ E K
Sbjct: 464 SPISVNFLTEITK 502
>TC8121 UP|Q9LKJ5 (Q9LKJ5) Multifunctional transport intrinsic membrane
protein 2, complete
Length = 1213
Score = 28.1 bits (61), Expect = 2.3
Identities = 15/49 (30%), Positives = 28/49 (56%)
Frame = +2
Query: 165 LKKRLSEIQWAAFILLTAGCTTAQLNSNSDHVLQTPFQGWVMAIAMALL 213
L+K ++E+ F + AGC + +N N+D+V+ P V +A+ +L
Sbjct: 209 LQKVIAELVGTYFFIF-AGCASIVVNKNNDNVVTLPGIALVWGLAVMVL 352
>TC12879
Length = 558
Score = 27.3 bits (59), Expect = 3.9
Identities = 17/52 (32%), Positives = 24/52 (45%), Gaps = 8/52 (15%)
Frame = +3
Query: 140 YVDAPGYQILKNFNIISTGV--------LYRIILKKRLSEIQWAAFILLTAG 183
YV G Q K +IS V L R ++++ LSE W F+L+ G
Sbjct: 6 YVQVNGRQYYKFVKVISGSVAKKNQTLPLPRSVVREYLSEHNWGRFVLMKEG 161
>TC17725 homologue to UP|Q8LLR2 (Q8LLR2) MADS-box protein 2, partial (45%)
Length = 970
Score = 26.6 bits (57), Expect = 6.6
Identities = 16/52 (30%), Positives = 30/52 (56%), Gaps = 4/52 (7%)
Frame = +3
Query: 50 FALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETL----KCAISLVALGR 97
+ L++L ++ LIV+S R YE+ +++ MV+TL KC+ V + +
Sbjct: 372 YELSVLCDAEVALIVFSTRGKLYEF--CSSSSMVKTLERYQKCSYGAVEVNK 521
>AU089094
Length = 639
Score = 26.6 bits (57), Expect = 6.6
Identities = 20/58 (34%), Positives = 31/58 (52%)
Frame = -2
Query: 37 RTQVQWKRKSMVTFALTILTSSQAILIVWSKRAGKYEYSVTTANFMVETLKCAISLVA 94
RT+V + + + L SS+AILI WS + YSV + + M++ L +SL A
Sbjct: 311 RTRVVERSRLCMADCKRSLQSSRAILIPWS----LFPYSVVSRSLMLKAL-MTLSLAA 153
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.137 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,236,884
Number of Sequences: 28460
Number of extensions: 62639
Number of successful extensions: 428
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 426
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 427
length of query: 347
length of database: 4,897,600
effective HSP length: 91
effective length of query: 256
effective length of database: 2,307,740
effective search space: 590781440
effective search space used: 590781440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0380.6


