

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0378b.5
(572 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
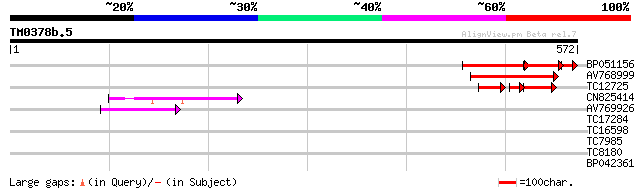
Score E
Sequences producing significant alignments: (bits) Value
BP051156 125 2e-49
AV768999 99 2e-21
TC12725 similar to GB|AAK32900.1|13605829|AF367313 AT5g04550/T32... 41 2e-10
CN825414 42 3e-04
AV769926 41 5e-04
TC17284 similar to UP|Q93WZ9 (Q93WZ9) Phosphoenolpyruvate carbox... 35 0.025
TC16598 weakly similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containi... 32 0.36
TC7985 similar to UP|O04002 (O04002) CDSP32 protein (Chloroplast... 28 5.3
TC8180 similar to UP|EXTN_TOBAC (P13983) Extensin precursor (Cel... 27 6.9
BP042361 27 6.9
>BP051156
Length = 500
Score = 125 bits (314), Expect(3) = 2e-49
Identities = 61/68 (89%), Positives = 63/68 (91%)
Frame = -3
Query: 457 KSSWVYNAEIAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATLKANILLVQTLYF 516
KSSWVYNAEI EWSAVLAQILDWLAPLAHDTIRW SVRSFEKEHATL+ANILLV TLYF
Sbjct: 498 KSSWVYNAEIPPEWSAVLAQILDWLAPLAHDTIRWPSVRSFEKEHATLEANILLVPTLYF 319
Query: 517 ANQVKTEA 524
AN VKTE+
Sbjct: 318 ANPVKTES 295
Score = 73.6 bits (179), Expect(3) = 2e-49
Identities = 35/40 (87%), Positives = 38/40 (94%)
Frame = -1
Query: 519 QVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVECASARSFH 558
Q+K AAIVDLLVGL+YVCRIDT+VGVRDRVECASARSFH
Sbjct: 311 QLKLRAAIVDLLVGLHYVCRIDTQVGVRDRVECASARSFH 192
Score = 35.4 bits (80), Expect(3) = 2e-49
Identities = 15/16 (93%), Positives = 15/16 (93%)
Frame = -2
Query: 557 FHGVGLRKNGMYSEYF 572
F GVGLRKNGMYSEYF
Sbjct: 196 FTGVGLRKNGMYSEYF 149
>AV768999
Length = 313
Score = 99.0 bits (245), Expect = 2e-21
Identities = 45/88 (51%), Positives = 64/88 (72%)
Frame = -1
Query: 466 IAAEWSAVLAQILDWLAPLAHDTIRWHSVRSFEKEHATLKANILLVQTLYFANQVKTEAA 525
+A EW L +IL WL+PLAH+ I+W S RSFE+++ K N+LL+QTL+FAN+ KTEAA
Sbjct: 304 LAGEWRDALGRILGWLSPLAHNMIKWQSERSFEQQNLVPKTNVLLLQTLFFANKEKTEAA 125
Query: 526 IVDLLVGLNYVCRIDTKVGVRDRVECAS 553
I +LLVGL Y+ R + ++ + ECA+
Sbjct: 124 ITELLVGLTYIWRFEREMTAKALFECAN 41
>TC12725 similar to GB|AAK32900.1|13605829|AF367313 AT5g04550/T32M21_140
{Arabidopsis thaliana;} , partial (11%)
Length = 425
Score = 41.2 bits (95), Expect(3) = 2e-10
Identities = 17/27 (62%), Positives = 21/27 (76%)
Frame = -2
Query: 474 LAQILDWLAPLAHDTIRWHSVRSFEKE 500
L +IL WL+PLAH+ IRW S RSFE +
Sbjct: 415 LGRILGWLSPLAHNMIRWQSERSFENQ 335
Score = 35.4 bits (80), Expect(3) = 2e-10
Identities = 16/33 (48%), Positives = 23/33 (69%)
Frame = -1
Query: 519 QVKTEAAIVDLLVGLNYVCRIDTKVGVRDRVEC 551
+ +TEAAI +LLVGLNYV R + ++ + EC
Sbjct: 278 RTQTEAAITELLVGLNYVWRFEREMTAKALFEC 180
Score = 24.6 bits (52), Expect(3) = 2e-10
Identities = 9/15 (60%), Positives = 14/15 (93%)
Frame = -3
Query: 505 KANILLVQTLYFANQ 519
+ N+LL+QTL+FA+Q
Sbjct: 321 RPNVLLLQTLFFADQ 277
>CN825414
Length = 597
Score = 42.0 bits (97), Expect = 3e-04
Identities = 34/146 (23%), Positives = 62/146 (42%), Gaps = 10/146 (6%)
Frame = +1
Query: 100 VARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGWDYRGKKMERK-----VKRMEKFVAAMT 154
V R G RC DP +H + YF+ G + +K ++ ++++ +V
Sbjct: 43 VVRFGNRCKDPQWHNLDR--------YFEKLGSELTPQKQLKEEAEVVMQQLMTYVQYTA 198
Query: 155 QLCQELEVLAEVEQTLRRM-----QANPVLHRGKLPEFQKKVTLQRQEVRNLRDMSPWNR 209
+L EL L +Q RR +N L + ++ Q++ VRNL+ S W++
Sbjct: 199 ELYHELHALDRFDQDYRRKLQEEDNSNATQRGDSLAILRSELKSQKKHVRNLKKKSLWSK 378
Query: 210 SYDYVVRLLVRSLFTILERIILVFGN 235
+ V+ V + + I FG+
Sbjct: 379 ILEEVMEKFVDVVNFLYLEIHEAFGS 456
>AV769926
Length = 280
Score = 41.2 bits (95), Expect = 5e-04
Identities = 24/82 (29%), Positives = 41/82 (49%), Gaps = 1/82 (1%)
Frame = +3
Query: 92 NFQSVARTVARLGRRCVDPVYHRFEHFVRSPAQNYFQWSGWDY-RGKKMERKVKRMEKFV 150
+ + A +V+RL R+C DP F A + W +++E K KRME++V
Sbjct: 27 SLRHTAESVSRLSRKCRDPALRSFPGVFAEFADSSLDPHAWTITTPREIEAKHKRMERYV 206
Query: 151 AAMTQLCQELEVLAEVEQTLRR 172
L +E++ L+ +E LR+
Sbjct: 207 TLTATLHREMDELSLLETALRK 272
>TC17284 similar to UP|Q93WZ9 (Q93WZ9) Phosphoenolpyruvate carboxylase
housekeeping isozyme pepc2 (Fragment) , partial (35%)
Length = 577
Score = 35.4 bits (80), Expect = 0.025
Identities = 22/68 (32%), Positives = 31/68 (45%), Gaps = 3/68 (4%)
Frame = -2
Query: 234 GNNHLATKQNENESPNTTAN---NLLRSQSFSVFMHSSVHPSENDLYGFHSGHLGRRLAS 290
GN+HL QN N + NTT N LR + + + +H S N L+G + G R
Sbjct: 516 GNSHLTLVQNSNVTTNTTHNIRETTLRMTLWQLCLTGVLHASSNTLHGQNKGVFKTRSIL 337
Query: 291 NSGILVDK 298
G+ K
Sbjct: 336 TFGVKFHK 313
>TC16598 weakly similar to UP|Q9SNC6 (Q9SNC6) Arm repeat containing protein
homolog, partial (15%)
Length = 589
Score = 31.6 bits (70), Expect = 0.36
Identities = 15/44 (34%), Positives = 24/44 (54%)
Frame = +2
Query: 11 WLSALWPVSRKGASDNKAVVVGVLGSEVAGLMLKVVNLWQSLSD 54
W LW S +G A + V+ E+AG ++++VN S+SD
Sbjct: 149 WRDLLWRASSRGGGGGGAAMEVVVEVEIAGSVIELVNEIASISD 280
>TC7985 similar to UP|O04002 (O04002) CDSP32 protein (Chloroplast
drought-induced stress protein of 32kDa), partial (80%)
Length = 1207
Score = 27.7 bits (60), Expect = 5.3
Identities = 20/64 (31%), Positives = 33/64 (51%)
Frame = +3
Query: 247 SPNTTANNLLRSQSFSVFMHSSVHPSENDLYGFHSGHLGRRLASNSGILVDKKQRKKKQQ 306
SP+T LLRS S S+ + + + L HS H+ + A++SG KK + ++
Sbjct: 114 SPSTHRKLLLRSSSLSITSTLTKNKTPIQLRASHS-HITKATAASSG---TKKAERDERV 281
Query: 307 QALH 310
Q +H
Sbjct: 282 QRVH 293
>TC8180 similar to UP|EXTN_TOBAC (P13983) Extensin precursor (Cell wall
hydroxyproline-rich glycoprotein), partial (3%)
Length = 575
Score = 27.3 bits (59), Expect = 6.9
Identities = 20/70 (28%), Positives = 31/70 (43%), Gaps = 2/70 (2%)
Frame = +2
Query: 108 VDPVYHRFEHFVRSPAQNYFQWS--GWDYRGKKMERKVKRMEKFVAAMTQLCQELEVLAE 165
VDP V+ +N WS G Y GKKME + K+ A + Q L+
Sbjct: 26 VDPALADLTESVKLSPRNVKAWSLLGECYEGKKMEEEAKK------AYQEALQLEPELSA 187
Query: 166 VEQTLRRMQA 175
++ L+R+ +
Sbjct: 188 AQEALKRLSS 217
>BP042361
Length = 394
Score = 27.3 bits (59), Expect = 6.9
Identities = 10/17 (58%), Positives = 15/17 (87%)
Frame = +3
Query: 296 VDKKQRKKKQQQALHPP 312
V+K+QR+++QQQ LH P
Sbjct: 300 VEKQQRRRQQQQQLHKP 350
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,187,534
Number of Sequences: 28460
Number of extensions: 114861
Number of successful extensions: 600
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 599
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 600
length of query: 572
length of database: 4,897,600
effective HSP length: 95
effective length of query: 477
effective length of database: 2,193,900
effective search space: 1046490300
effective search space used: 1046490300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0378b.5


