

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371b.6
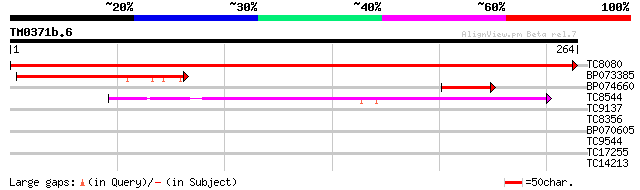
(264 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8080 536 e-153
BP073385 126 4e-30
BP074660 57 2e-09
TC8544 47 3e-06
TC9137 UP|CAF02297 (CAF02297) Cysteine-rich polycomb-like protei... 27 2.8
TC8356 similar to GB|AAM64696.1|21592747|AY087138 gamma-tocopher... 27 4.7
BP070605 26 8.0
TC9544 UP|AAO92441 (AAO92441) Homoglutathione synthetase, complete 26 8.0
TC17255 26 8.0
TC14213 26 8.0
>TC8080
Length = 1432
Score = 536 bits (1382), Expect = e-153
Identities = 263/264 (99%), Positives = 264/264 (99%)
Frame = +1
Query: 1 MDTTNDWVLLSDDGSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHD 60
MDTTNDWVLLSDDGSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHD
Sbjct: 163 MDTTNDWVLLSDDGSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQLLHD 342
Query: 61 DPIQFVENTEDDHVGDNSDTTSEKIKAVVVEADEQETLSVSQFFLQLENKFADMKMDSPK 120
DPIQFVE+TEDDHVGDNSDTTSEKIKAVVVEADEQETLSVSQFFLQLENKFADMKMDSPK
Sbjct: 343 DPIQFVEHTEDDHVGDNSDTTSEKIKAVVVEADEQETLSVSQFFLQLENKFADMKMDSPK 522
Query: 121 SSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKW 180
SSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKW
Sbjct: 523 SSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATMDCDREEVDPTDGFNFWKW 702
Query: 181 SLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQATKLNE 240
SLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQATKLNE
Sbjct: 703 SLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQIYTDDKRIKQVVQQATKLNE 882
Query: 241 AIAATRGIPLSRAHITYGGYYEAL 264
AIAATRGIPLSRAHITYGGYYEAL
Sbjct: 883 AIAATRGIPLSRAHITYGGYYEAL 954
>BP073385
Length = 519
Score = 126 bits (316), Expect = 4e-30
Identities = 70/92 (76%), Positives = 73/92 (79%), Gaps = 12/92 (13%)
Frame = +1
Query: 4 TNDWVLLSDDGSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSS--TTQLLHDD 61
+ DWVLLSDDGSLDGGDENQI L KSNPES+SD DMNYFYCTSPKSRDSS TTQLLHDD
Sbjct: 244 SRDWVLLSDDGSLDGGDENQICL*KSNPESESDLDMNYFYCTSPKSRDSSTTTTQLLHDD 423
Query: 62 PIQF--------VENTE-DDHVGDNS-DTTSE 83
PI VE TE DDHVGDNS DT+SE
Sbjct: 424 PILLSVPHIGNKVEKTEDDDHVGDNSADTSSE 519
>BP074660
Length = 386
Score = 57.4 bits (137), Expect = 2e-09
Identities = 25/25 (100%), Positives = 25/25 (100%)
Frame = -1
Query: 202 VFGSQQKNKFHQNHKIQFQIYTDDK 226
VFGSQQKNKFHQNHKIQFQIYTDDK
Sbjct: 386 VFGSQQKNKFHQNHKIQFQIYTDDK 312
>TC8544
Length = 1278
Score = 47.0 bits (110), Expect = 3e-06
Identities = 46/212 (21%), Positives = 90/212 (41%), Gaps = 6/212 (2%)
Frame = +2
Query: 47 PKSRDSSTTQLLHDDPIQFVENTEDDHVGDNSDTTSEKIKAVVVEADEQETLSVSQFFLQ 106
P D++ L+ D +++T + + NS+++S++ V+ + ++ +Q F
Sbjct: 188 PVVSDATNLALIRPDHFS-IDSTPNSNSSCNSNSSSDE-----VDRNFDDSFVANQLFPN 349
Query: 107 LENKFADMKMDSPKSSPRVLFPPPLDAGALKFEDKGEAMEVMVSPRRKVENEMATM---D 163
+ DS + + A A + E +EV V KVE E +
Sbjct: 350 PNPQRWTHDSDSDSITTDPVKETEDSASAAAASSESEEVEVGVGFDSKVEGEEVKAIEKE 529
Query: 164 CDREEV---DPTDGFNFWKWSLTGVGAICSFGVAAATICVLVFGSQQKNKFHQNHKIQFQ 220
DR V P + +W + +CS AAA + +L G + + ++
Sbjct: 530 EDRRVVWWRVPFEVLRYWVNPIPLSLPLCSVAAAAAFLGLLFLGRRLYKMKRKTQTMKLN 709
Query: 221 IYTDDKRIKQVVQQATKLNEAIAATRGIPLSR 252
+ DDK++ Q++ + +LNEA + R +P+ R
Sbjct: 710 LALDDKKVSQLMVRVARLNEAFSVVRRVPVVR 805
>TC9137 UP|CAF02297 (CAF02297) Cysteine-rich polycomb-like protein,
complete
Length = 3086
Score = 27.3 bits (59), Expect = 2.8
Identities = 14/41 (34%), Positives = 24/41 (58%), Gaps = 1/41 (2%)
Frame = +1
Query: 205 SQQKNKFHQNHKIQFQIYTDDKR-IKQVVQQATKLNEAIAA 244
S++ N+ HQ ++ DKR +++V Q+ TKL +AA
Sbjct: 697 SEESNEIHQERPVENPETEGDKRVVERVSQEHTKLESNLAA 819
>TC8356 similar to GB|AAM64696.1|21592747|AY087138 gamma-tocopherol
methyltransferase {Arabidopsis thaliana;} , partial
(30%)
Length = 642
Score = 26.6 bits (57), Expect = 4.7
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Frame = +1
Query: 185 VGAICSFGVAAATICVLVFGSQ-QKNKFHQNHKIQFQIYTDDKRIKQVVQQ 234
V CS + T+ + + GS ++++NHK F DD+RI++ Q
Sbjct: 139 VSLCCSILASCDTLSIYMEGSHFTVAQWNENHKRSFGYAIDDRRIQEGCHQ 291
>BP070605
Length = 393
Score = 25.8 bits (55), Expect = 8.0
Identities = 14/36 (38%), Positives = 19/36 (51%), Gaps = 6/36 (16%)
Frame = -3
Query: 100 VSQFFLQLENKFADMKMDSP------KSSPRVLFPP 129
+S+F LQL + + MDSP SP V+ PP
Sbjct: 370 ISRFLLQLGMRHSQHHMDSPHHSQHHTGSPTVIHPP 263
>TC9544 UP|AAO92441 (AAO92441) Homoglutathione synthetase, complete
Length = 1860
Score = 25.8 bits (55), Expect = 8.0
Identities = 12/23 (52%), Positives = 14/23 (60%)
Frame = -3
Query: 108 ENKFADMKMDSPKSSPRVLFPPP 130
E F + S KSSP +LFPPP
Sbjct: 1393 EPDFCNFMRVSLKSSP*ILFPPP 1325
>TC17255
Length = 485
Score = 25.8 bits (55), Expect = 8.0
Identities = 19/57 (33%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Frame = +1
Query: 2 DTTN-DWVLLSDDGSLDGGDENQIFLGKSNPESKSDFDMNYFYCTSPKSRDSSTTQL 57
DT N D V+ +DD L D++ E KS ++YF PK+R S Q+
Sbjct: 103 DTMNIDAVIRTDDAELGEADQD---------EPKSGDPLDYFLPPPPKARCSEELQV 246
>TC14213
Length = 1284
Score = 25.8 bits (55), Expect = 8.0
Identities = 14/36 (38%), Positives = 19/36 (51%), Gaps = 6/36 (16%)
Frame = +2
Query: 100 VSQFFLQLENKFADMKMDSP------KSSPRVLFPP 129
+S+F LQL + + MDSP SP V+ PP
Sbjct: 782 ISRFLLQLGMRHSQHHMDSPHHSQHHTGSPTVIHPP 889
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,224,354
Number of Sequences: 28460
Number of extensions: 53858
Number of successful extensions: 310
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 308
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 309
length of query: 264
length of database: 4,897,600
effective HSP length: 88
effective length of query: 176
effective length of database: 2,393,120
effective search space: 421189120
effective search space used: 421189120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0371b.6


