

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0368c.9
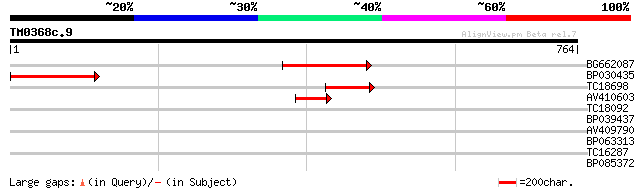
(764 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG662087 207 4e-54
BP030435 162 2e-40
TC18698 72 3e-13
AV410603 51 8e-07
TC18092 35 0.059
BP039437 28 5.5
AV409790 28 5.5
BP063313 28 7.2
TC16287 homologue to BAD12186 (BAD12186) 12-oxophytodienoic acid... 28 7.2
BP085372 27 9.4
>BG662087
Length = 373
Score = 207 bits (528), Expect = 4e-54
Identities = 101/120 (84%), Positives = 107/120 (89%)
Frame = +1
Query: 368 NGK*PMCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKD 427
+GK M VDYTDLNKACPKDSYPLPSIDKLVDGAS+NELLSLMDAYSGYHQIKMH SD+D
Sbjct: 13 SGKWRMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDED 192
Query: 428 KTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEANH 487
KTAFMTAR NY YQT+PFGLKN GATYQ LMDRVF D VGRNMEVY+ +MIVKS L ANH
Sbjct: 193 KTAFMTARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIVKSALRANH 372
>BP030435
Length = 533
Score = 162 bits (409), Expect = 2e-40
Identities = 81/120 (67%), Positives = 93/120 (77%)
Frame = +1
Query: 1 LLAARRPMSARSGKNSMPKEVGAIAGGFGGGGVTSAARKRYVRAVNAVTEVPFGFLHPDI 60
L+A +P + G +PK V IAGGFGGGG+TSAARKRY RAVN VTEV FGF HP I
Sbjct: 172 LIATGKPTRIQGGNAPVPKGVHTIAGGFGGGGITSAARKRYARAVNTVTEVLFGFSHPVI 351
Query: 61 TISSADFFGVEQHLYDPIVVLLRINQFNVQRVLLDQGSSADIIYGDAFDQLGLDERDLTP 120
T SS DF ++ HL +PIV+LLR+NQ NVQRVLLDQGSSADIIYG FD+LGL+E DLTP
Sbjct: 352 TFSSDDFIRIKPHLDNPIVILLRVNQLNVQRVLLDQGSSADIIYGGVFDRLGLNEADLTP 531
>TC18698
Length = 808
Score = 72.4 bits (176), Expect = 3e-13
Identities = 37/66 (56%), Positives = 43/66 (65%)
Frame = -2
Query: 426 KDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFADQVGRNMEVYVGDMIVKSVLEA 485
K KT R NY YQ MP GLKN TYQRLMD++F Q+ +N+EVYV DMIVKS E
Sbjct: 807 KKKTTLKINRVNYYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIVKSSQE* 628
Query: 486 NHHEEL 491
H +L
Sbjct: 627 FHRGDL 610
>AV410603
Length = 162
Score = 50.8 bits (120), Expect = 8e-07
Identities = 21/48 (43%), Positives = 32/48 (65%)
Frame = +1
Query: 386 KDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQIKMHSSDKDKTAFMT 433
KDS+P+P++D+L+D ++ S +D SGYHQI + D+ KT F T
Sbjct: 10 KDSFPMPTVDELLDELRGSQFFSKLDLRSGYHQILVKPEDRHKTVFRT 153
>TC18092
Length = 519
Score = 34.7 bits (78), Expect = 0.059
Identities = 19/35 (54%), Positives = 24/35 (68%)
Frame = -2
Query: 373 MCVDYTDLNKACPKDSYPLPSIDKLVDGASNNELL 407
MCV TD+NK CP+ PL SI+ LVD +S+ E L
Sbjct: 101 MCVC*TDINKDCPR--XPLTSINALVDYSSSYEYL 3
>BP039437
Length = 524
Score = 28.1 bits (61), Expect = 5.5
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 1/80 (1%)
Frame = -2
Query: 409 LMDAYSGYHQIKMHSSDKDKTAFMTARANYSYQTMPFGLKNTGATYQRLMDRVFA-DQVG 467
L + +S ++ + S D D +T+ YS + M +KN + FA +VG
Sbjct: 493 LHELFSNGIRVWIFSGDTDGRVPVTS-TKYSIKKMNLPIKNVWRPW-------FAYGEVG 338
Query: 468 RNMEVYVGDMIVKSVLEANH 487
EVY GD+ +V EA H
Sbjct: 337 GYTEVYKGDLTFATVREAGH 278
>AV409790
Length = 421
Score = 28.1 bits (61), Expect = 5.5
Identities = 21/52 (40%), Positives = 25/52 (47%), Gaps = 6/52 (11%)
Frame = -3
Query: 692 GRC------CRSQTYQEGSSRGQ*TCRSMVCSMTREGR*ERKL*QISW*S*H 737
GRC RS+ +QEGS R CR+ +CS GR* W* *H
Sbjct: 392 GRCGKWRWGARSRCFQEGSCRHLRECRATLCSHIC*GR*R-------W*L*H 258
>BP063313
Length = 386
Score = 27.7 bits (60), Expect = 7.2
Identities = 12/35 (34%), Positives = 19/35 (54%)
Frame = +3
Query: 211 KCYCNAIDRYGRKSALTGHRCHEVNVPEENLDSRG 245
K Y IDR+G++ AL + EEN+++ G
Sbjct: 102 KAYIRPIDRFGQRYALDPFFTKLTGITEENIETEG 206
>TC16287 homologue to BAD12186 (BAD12186) 12-oxophytodienoic acid
10,11-reductase, partial (23%)
Length = 534
Score = 27.7 bits (60), Expect = 7.2
Identities = 13/38 (34%), Positives = 23/38 (60%)
Frame = -1
Query: 381 NKACPKDSYPLPSIDKLVDGASNNELLSLMDAYSGYHQ 418
NK CP SY SID ++ +++ S ++++S +HQ
Sbjct: 132 NKVCPVFSY---SIDPILSSITSSNYKSSIESFSHWHQ 28
>BP085372
Length = 459
Score = 27.3 bits (59), Expect = 9.4
Identities = 12/24 (50%), Positives = 14/24 (58%)
Frame = -3
Query: 13 GKNSMPKEVGAIAGGFGGGGVTSA 36
G +P G GG+GG GVTSA
Sbjct: 403 GYGGVPGSYGGGGGGYGGQGVTSA 332
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.337 0.145 0.453
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,377,842
Number of Sequences: 28460
Number of extensions: 137949
Number of successful extensions: 1016
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 998
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1015
length of query: 764
length of database: 4,897,600
effective HSP length: 97
effective length of query: 667
effective length of database: 2,136,980
effective search space: 1425365660
effective search space used: 1425365660
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0368c.9


