

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
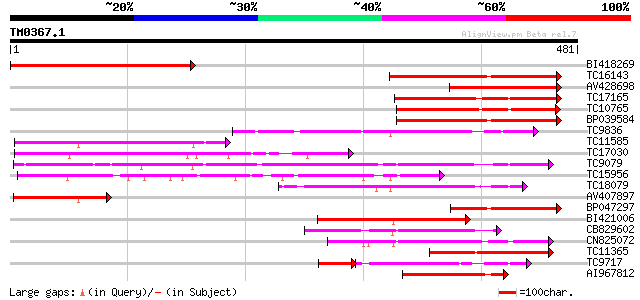
Query= TM0367.1
(481 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI418269 324 2e-89
TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltrans... 196 5e-51
AV428698 186 6e-48
TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltran... 151 2e-37
TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid gl... 149 1e-36
BP039584 144 4e-35
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 141 2e-34
TC11585 similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid glyc... 137 4e-33
TC17030 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, pa... 131 2e-31
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 130 6e-31
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 129 1e-30
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 122 1e-28
AV407897 118 2e-27
BP047297 118 2e-27
BI421006 110 7e-25
CB829602 107 4e-24
CN825072 105 2e-23
TC11365 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, pa... 98 3e-21
TC9717 weakly similar to UP|Q9LNI1 (Q9LNI1) F6F3.22 protein, par... 80 4e-20
AI967812 94 6e-20
>BI418269
Length = 520
Score = 324 bits (830), Expect = 2e-89
Identities = 157/157 (100%), Positives = 157/157 (100%)
Frame = +2
Query: 1 MDVESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLH 60
MDVESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLH
Sbjct: 50 MDVESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLH 229
Query: 61 LHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYC 120
LHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYC
Sbjct: 230 LHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVADYC 409
Query: 121 FPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSS 157
FPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSS
Sbjct: 410 FPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRGSSS 520
>TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (30%)
Length = 604
Score = 196 bits (499), Expect = 5e-51
Identities = 100/147 (68%), Positives = 120/147 (81%), Gaps = 1/147 (0%)
Frame = +1
Query: 323 EKGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMT 382
EKGMI+RGW PQ++ILGH +VGAF+THCG NS +EAVSAGVP+ITWPV+G+QFY EKL+T
Sbjct: 4 EKGMIIRGWVPQVVILGHRAVGAFVTHCGWNSTVEAVSAGVPMITWPVVGEQFYNEKLIT 183
Query: 383 EVRGVGVEVGAEEWS-MGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGN 441
EVRG+GVEVGAEE MG +RE+ ++SR+ IEKAVRRLM GG+E EQIRRR E G
Sbjct: 184 EVRGIGVEVGAEECCLMGFDKREK---LVSRESIEKAVRRLMDGGDEGEQIRRRAQEYGE 354
Query: 442 LANAAVQPGGSSYQNLTALIAHLKTLR 468
A AV+ GGSS++NLTALI LK LR
Sbjct: 355 KARQAVEEGGSSHKNLTALIDDLKRLR 435
>AV428698
Length = 287
Score = 186 bits (473), Expect = 6e-48
Identities = 94/95 (98%), Positives = 95/95 (99%)
Frame = -1
Query: 374 QFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIR 433
QFYTEKLMTEVRGVGV+VGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIR
Sbjct: 287 QFYTEKLMTEVRGVGVKVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIR 108
Query: 434 RRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLR 468
RRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLR
Sbjct: 107 RRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLR 3
>TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltransferase,
partial (24%)
Length = 546
Score = 151 bits (382), Expect = 2e-37
Identities = 78/142 (54%), Positives = 100/142 (69%)
Frame = +1
Query: 327 ILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRG 386
I+RGWAPQ+LIL H ++GAF+THCG NS LE V+AGVP++TWP+ +QFY EKL+TEV
Sbjct: 1 IIRGWAPQVLILEHEAIGAFVTHCGWNSTLEGVAAGVPMVTWPIAAEQFYNEKLVTEVLK 180
Query: 387 VGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAA 446
+GV VGA++W GR E + D +EK V+R+M GEEAE++R RV L A A
Sbjct: 181 IGVPVGAKKW----GRMVLEGDGVKSDAVEKGVKRIM-EGEEAEEMRNRVKVLSQQAQWA 345
Query: 447 VQPGGSSYQNLTALIAHLKTLR 468
V+ GGSSY +L ALI L LR
Sbjct: 346 VEEGGSSYSDLNALIEELGMLR 411
>TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (19%)
Length = 542
Score = 149 bits (375), Expect = 1e-36
Identities = 78/139 (56%), Positives = 100/139 (71%)
Frame = +2
Query: 329 RGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVG 388
+GWAPQ LIL H + G FLTHCG N+ +EA+SAGVP+IT P DQ+Y EKL+TEV G G
Sbjct: 2 KGWAPQPLILNHLATGGFLTHCGWNAVVEAISAGVPMITMPGFSDQYYNEKLITEVHGFG 181
Query: 389 VEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQ 448
VEVGA EWS+ + ++ V+S ++IEKAV+ LM GEE +IR + E+ + A AVQ
Sbjct: 182 VEVGAAEWSI--SPYDGKKKVVSGERIEKAVKSLM--GEEGAEIRSKAKEVQDKAWKAVQ 349
Query: 449 PGGSSYQNLTALIAHLKTL 467
GGSSY +LT LI HL+TL
Sbjct: 350 QGGSSYNSLTVLIDHLRTL 406
>BP039584
Length = 545
Score = 144 bits (362), Expect = 4e-35
Identities = 69/140 (49%), Positives = 94/140 (66%)
Frame = -3
Query: 329 RGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVG 388
RGWAPQLLIL HP++G +THCG N+ LE+V AG+P+ T P+ +QFY EKL+ +V GVG
Sbjct: 543 RGWAPQLLILEHPAIGGVVTHCGWNTTLESVIAGLPMATMPLFAEQFYNEKLLVDVLGVG 364
Query: 389 VEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNLANAAVQ 448
V +G ++W + ++ R+ I KA+ LMGGGEEA ++RRRV EL + A +Q
Sbjct: 363 VSIGMKKWRNWNVVGDE---IVKRENIVKAISLLMGGGEEALEMRRRVRELSDAAKKTIQ 193
Query: 449 PGGSSYQNLTALIAHLKTLR 468
PGGSSY + L LK L+
Sbjct: 192 PGGSSYNKVKGLFDELKALK 133
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 141 bits (356), Expect = 2e-34
Identities = 90/263 (34%), Positives = 132/263 (49%), Gaps = 4/263 (1%)
Frame = +1
Query: 190 LKSHGVLINSFVELDGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVST 249
+ S G L N+ + D Y AW +GP L G+ G +S
Sbjct: 214 VNSDGFLFNTVADFDSMGL--RYVARKLNRPAWAIGPVLLSTGSGSRGKGGG-----ISP 372
Query: 250 DELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEE 309
+ WL TK +SVL+V FGSM + S Q+ ++A ++ S F+WVV G +
Sbjct: 373 ELCRKWLDTKPSNSVLFVSFGSMNTISASQMMQLATALDRSGRNFIWVV-RPPIGFDINS 549
Query: 310 EEEKEKWLPEGF----EEKGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPI 365
E E+WLPEGF +KG+++ WAPQ+ IL H +V AFL+HCG NS LE++S GVPI
Sbjct: 550 EFRAEEWLPEGFLGRVAKKGLVVHDWAPQVEILSHGAVSAFLSHCGWNSVLESLSHGVPI 729
Query: 366 ITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGG 425
+ WP+ +QF+ K++ E GV VEV R + V D +EK + +M
Sbjct: 730 LGWPMAAEQFFNCKMLEEELGVCVEVA----------RGKRCEVRHEDLVEK-IELVMNE 876
Query: 426 GEEAEQIRRRVHELGNLANAAVQ 448
E +IR+ + + AV+
Sbjct: 877 AESGVKIRKNAGNIREMIRDAVR 945
>TC11585 similar to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (40%)
Length = 609
Score = 137 bits (345), Expect = 4e-33
Identities = 75/199 (37%), Positives = 108/199 (53%), Gaps = 16/199 (8%)
Frame = +1
Query: 5 SEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLP------- 57
SE LK+YF+P+ A GH+IPL +A L ASRG+HVTIITTP+N D ++
Sbjct: 13 SEMETLKVYFVPFFAQGHLIPLVHLARLVASRGEHVTIITTPANAQLFDKTIEEDRACGY 192
Query: 58 NLHLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPIQLFMEHHPADCVVA 117
++ +H V FPS QVGLP G+E+ +++D T K+H L++ I+ FM+ P D ++
Sbjct: 193 HIRVHLVKFPSTQVGLPGGVENLFAASDNLTAGKIHMAAHLIQPEIEGFMKQSPPDVLIP 372
Query: 118 DYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKRG---------SSSFLISGLPHGDI 168
D F W + L IPRL FNP +F +C + A + S + I GLPH +
Sbjct: 373 DIMFTWSEASAKSLGIPRLIFNPISIFDVCMIEAIKSHPEAFTCSDSGPYHIPGLPH-SL 549
Query: 169 ALNASPPEALTACVEPLLR 187
L P E L++
Sbjct: 550 TLPIKPSPGFARLTESLVQ 606
>TC17030 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (51%)
Length = 1098
Score = 131 bits (330), Expect = 2e-31
Identities = 96/313 (30%), Positives = 148/313 (46%), Gaps = 26/313 (8%)
Frame = +1
Query: 5 SEQRPLKLYFIPYLAAGHMIPLCDIA-ILFASRGQHVTIITTPSNT---TTIDSSLPNLH 60
S+ + P ++ GHMIP+ DIA IL + VT+ITTP N T S +
Sbjct: 193 SQASQIHFVLFPLMSQGHMIPMMDIATILSQQQDVTVTVITTPHNASRFTHTTKSRSQIR 372
Query: 61 LHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLLREPI-QLFME-HHPADCVVAD 118
+ + FP + GLP+G E+ L T L++P+ QLF E P +C+++D
Sbjct: 373 ILELEFPHNETGLPEGCENLDMLPSLGTALTFFNAASNLQKPVEQLFNELTPPPNCIISD 552
Query: 119 YCFPWIDDLTTELHIPRLSFNPSPLFALCAM------RAKRGSSS----FLISGLPHGDI 168
C P+ + + +IPR+SF F L + + ++ +S F++ G+P
Sbjct: 553 MCLPYTARIAAKFNIPRISFLGQSCFTLFCLYNIGRHKVRQNVTSETEYFVLPGVPDKIE 732
Query: 169 ALNASPPEALTACVEP--------LLRKELKSHGVLINSFVELDGHEYVKHYERTTGGHK 220
A P A ++ E S+GV++NSF EL+ EY K Y++ G K
Sbjct: 733 MTKAQIPGPSGARMDQNRIAFYAETAAAEAASYGVVMNSFEELET-EYAKGYKKVKNG-K 906
Query: 221 AWLLGPASLVRGTAKEKAERGHEKSVVSTDE--LLSWLSTKKPSSVLYVCFGSMCSFSNE 278
W +GP SL + K + DE + WL +K SV+Y C GSMC+ +
Sbjct: 907 VWCIGPVSL----------SNNNKVSIDEDEHYCMKWLDLQKSKSVIYACLGSMCNLTPL 1056
Query: 279 QLHEMACGIEASS 291
QL E+ G+EAS+
Sbjct: 1057QLIELGLGLEASN 1095
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 130 bits (326), Expect = 6e-31
Identities = 123/499 (24%), Positives = 204/499 (40%), Gaps = 41/499 (8%)
Frame = +1
Query: 4 ESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLPNLHLHT 63
++E++P + P GH+ PL +A L RG H+T + T N + S L
Sbjct: 1 QAERKPHAV-LTPSPLQGHINPLFQLAKLLHLRGFHITFVHTEYNHKRLIKSRGPNALDG 177
Query: 64 VPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIK-LLREPIQ-LFMEHH---------PA 112
+P + +PDG+E Q + + I+ P Q L + H P
Sbjct: 178 LPDFVFET-IPDGLEDGDGEVS-QDLYSLCDSIRNKCHLPFQDLLAKLHRSASAGLVPPV 351
Query: 113 DCVVADYCFPWIDDLTTELHIPRLSFNPSPLFALCAMRAKR------------------- 153
C+V+D + +L +P L P+ + +
Sbjct: 352 TCLVSDLLMTFTIQAAQQLQLPILLLCPASASTFMSFLHFQTLLDKGVIPLKDESYLTNG 531
Query: 154 ----------GSSSFLISGLPHGDIALNASPPEALTACVEPLLRKELKSHGVLINSFVEL 203
G +F + LP D P + + + + + ++ ++ N+F EL
Sbjct: 532 YLDTKVDWIPGMQNFRLKDLP--DFIRTTDPNDIMLEFMVEVADRAKRASAIVFNTFNEL 705
Query: 204 DGHEYVKHYERTTGGHKAWLLGPASLVRGTAKEKAERGHEKSVVSTD-ELLSWLSTKKPS 262
+ +T + +GP A + ++ D + L WL +K+P
Sbjct: 706 ERDVLSA---LSTMLPSLYPIGPFPSFLNQAPQNQLASLGSNLWKEDTKCLQWLESKEPG 876
Query: 263 SVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEGFE 322
SV+YV FGS+ S EQL E A G+ F+W++ + K +
Sbjct: 877 SVVYVNFGSITVMSPEQLLEFAWGLANGKKPFLWIIRPDLVADGSAILSPK---FVDETS 1047
Query: 323 EKGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMT 382
+G+I W PQ +L HPSVG FLTHCG NS +E++ AGVP++ WP + DQ + +
Sbjct: 1048NRGLIA-SWCPQEKVLNHPSVGGFLTHCGWNSTIESICAGVPMLCWPFVADQPTNCRYIC 1224
Query: 383 EVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHELGNL 442
+G+EV + R+++E + LM G++ +++R+R EL
Sbjct: 1225NEWDIGIEVDTN---------------VKREEVENLIHELM-VGDKGKKMRQRTMELKKK 1356
Query: 443 ANAAVQPGGSSYQNLTALI 461
A +PGG SY NL LI
Sbjct: 1357AEEDTRPGGCSYMNLDKLI 1413
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 129 bits (323), Expect = 1e-30
Identities = 114/404 (28%), Positives = 196/404 (48%), Gaps = 41/404 (10%)
Frame = +2
Query: 7 QRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPS------NTTTIDSSLPN-L 59
+ PL + + + A GH+ PL +A A++G VT+ TT + T I P +
Sbjct: 26 ESPLHILLVSFPAQGHINPLLRLAKCLAAKGSSVTLATTQTAGKSMRTATNITDKAPTPI 205
Query: 60 HLHTVPFPSRQVGLPDGIESFSSSADLQTTFKVHQGIKLL--REPIQLFMEHHPAD---- 113
++ F GL +S + L Q ++L+ ++ +++ EH ++
Sbjct: 206 SDGSLKFEFFDDGLGPDDDSIRGNLSLYL-----QQLELVGRQQILKMIKEHADSNRQIS 370
Query: 114 CVVADYCFPWIDDLTTELHIPR--LSFNPSPLFA--------LCAMRAKRGSSSFLISGL 163
C++ + PW+ D+ E IP L S +F L +K + + L
Sbjct: 371 CIINNPFLPWVVDVAEEQGIPSALLWIQSSAVFTAYYSYFHNLVTFPSKEDPLADV--QL 544
Query: 164 PHGDIAL-NASPPEAL-TACVEPLLRKEL--------KSHGVLINSFVELDGHEYVKHYE 213
P I L + P+ L +C P L + K+ +L++++ EL+ H+++ +
Sbjct: 545 PSTSIVLKHCEIPDFLHPSCTYPFLGTLILEQFKNLNKTFCILVDTYEELE-HDFIDYLS 721
Query: 214 RTTGGHKAWLLGPASLV--RGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGS 271
+T +L+ P + AK A RG + +D+ + WL++K SSV+YV FGS
Sbjct: 722 KT------FLIRPVGPLFKSSQAKSAAIRG---DFMKSDDCIEWLNSKPQSSVVYVSFGS 874
Query: 272 MCSFSNEQLHEMACGIEASSVEFVWVV--PAEKRGKEEEEEEEKEKWLPEGF----EEKG 325
+ EQ+ E+A G++ S V F+WV+ P E G + LP GF E+G
Sbjct: 875 IVYLPQEQVDEIAFGLKKSQVSFLWVMKPPPEVTGLQPHV-------LPYGFLEETGERG 1033
Query: 326 MILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWP 369
+++ W+PQ +L HPSV FLTHCG NS++EA+++GVP++T+P
Sbjct: 1034RVVQ-WSPQEEVLAHPSVSCFLTHCGWNSSMEALTSGVPVLTFP 1162
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 122 bits (307), Expect = 1e-28
Identities = 83/220 (37%), Positives = 113/220 (50%), Gaps = 9/220 (4%)
Frame = +3
Query: 229 LVRGTAKEKAERGHEKSVVSTDELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIE 288
LVR T + K G ++ DE+L WL + SV+YV FGS + Q+ E+A G+E
Sbjct: 9 LVR-TVESKPRHGEDQ-----DEILRWLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGLE 170
Query: 289 ASSVEFVWVVPAEKRGKEEEE-----EEEKEKWLPEGF----EEKGMILRGWAPQLLILG 339
S FVWVV E +LPEGF E G+++ WAPQ ILG
Sbjct: 171 LSQHRFVWVVRPPNEADASATFFGFGNEMALNYLPEGFMNRTREFGVVVPMWAPQAEILG 350
Query: 340 HPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMG 399
HP+ G F+T CG NS LE+V GVP++ WP+ +Q +++E GV V V E G
Sbjct: 351 HPATGGFVTPCGWNSVLESVLNGVPMVAWPLYSEQKMNAYMLSEELGVAVRVKVAE---G 521
Query: 400 GGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHEL 439
G V+ RD++ VRR+M EE ++ RV EL
Sbjct: 522 G--------VVCRDQVVDMVRRVM-VSEEGMGMKDRVREL 614
>AV407897
Length = 295
Score = 118 bits (296), Expect = 2e-27
Identities = 57/85 (67%), Positives = 68/85 (79%), Gaps = 2/85 (2%)
Frame = +2
Query: 4 ESEQRPLKLYFIPYLAAGHMIPLCDIAILFASRGQHVTIITTPSNTTTIDSSLP--NLHL 61
E E++PLKLYFI YLAAGHMIPLCDIA LFASRG HVTIITTPSN + S+P NL L
Sbjct: 38 EPEEKPLKLYFIHYLAAGHMIPLCDIATLFASRGHHVTIITTPSNAQILQKSIPSHNLKL 217
Query: 62 HTVPFPSRQVGLPDGIESFSSSADL 86
H V FP++++GLPDG+ES S+ D+
Sbjct: 218 HAVKFPAQELGLPDGVESLSAVNDI 292
>BP047297
Length = 490
Score = 118 bits (295), Expect = 2e-27
Identities = 63/95 (66%), Positives = 74/95 (77%), Gaps = 1/95 (1%)
Frame = -1
Query: 375 FYTEKLMTEVRGVGVEVGAEEWS-MGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIR 433
FY EKL+TEVRG+GVEVGAEEW+ +G G RE+ V+SR+ IEKAVRRLM GG+EAEQIR
Sbjct: 490 FYNEKLITEVRGIGVEVGAEEWTPIGFGEREK---VVSRESIEKAVRRLMDGGDEAEQIR 320
Query: 434 RRVHELGNLANAAVQPGGSSYQNLTALIAHLKTLR 468
RR HE G A AV+ GGSS+ LTALI +K LR
Sbjct: 319 RRAHEYGRKARLAVEEGGSSHNXLTALIDDIKRLR 215
>BI421006
Length = 519
Score = 110 bits (274), Expect = 7e-25
Identities = 55/135 (40%), Positives = 85/135 (62%), Gaps = 5/135 (3%)
Frame = +2
Query: 262 SSVLYVC-FGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEEEEKEKWLPEG 320
+ + +C F +F EQ+ ++A G+E S +F+WV+ +G + ++ KE+ LP+G
Sbjct: 77 AKICSICIFWFFSTFIEEQIEQIANGLEQSKQKFIWVLRDADKGDIFDGDKVKERGLPKG 256
Query: 321 FEEK----GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFY 376
FE++ G+++R WAPQL IL HPS G F++HCG NS +E++S GVPI WP+ DQ
Sbjct: 257 FEKRVEGMGLVVRDWAPQLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPR 436
Query: 377 TEKLMTEVRGVGVEV 391
L+TEV V + V
Sbjct: 437 NTVLITEVLKVALVV 481
>CB829602
Length = 542
Score = 107 bits (267), Expect = 4e-24
Identities = 62/170 (36%), Positives = 96/170 (56%), Gaps = 3/170 (1%)
Frame = +1
Query: 251 ELLSWLSTKKPSSVLYVCFGSMCSFSNEQLHEMACGIEASSVEFVWVVPAEKRGKEEEEE 310
E L WL +++PSSVLYV FGS+ + + +QL E+A GI S +F+WV+ + + E
Sbjct: 91 ECLKWLDSQEPSSVLYVNFGSVINMTPQQLVELAWGIANSKKKFIWVI------RPDLVE 252
Query: 311 EEKEKWLPEGFEE---KGMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIIT 367
E LPE E +G++L W PQ +L H ++G FLTHCG NS +E++S+GVP+I
Sbjct: 253 GEASIVLPEIVAETKDRGIML-SWCPQEQVLKHSALGGFLTHCGWNSTIESISSGVPLIC 429
Query: 368 WPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIEK 417
P DQF + + G+E+ +++ + RD++EK
Sbjct: 430 SPFFNDQFVNSRYICSEWDFGMEMNSDD--------------VKRDEVEK 537
>CN825072
Length = 712
Score = 105 bits (262), Expect = 2e-23
Identities = 67/202 (33%), Positives = 106/202 (52%), Gaps = 10/202 (4%)
Frame = -3
Query: 270 GSMCSFSNEQLHEMACGIEASSVEFVWVV--PAEKR---GKEEEEEEEKEKWLPEGFEEK 324
GS SF Q+ E+A +E S V FVW + P K G + ++ LPEGF ++
Sbjct: 710 GSRGSFDRAQITEIARAVENSGVRFVWSLRKPXLKGSMVGPSDYSVDDLASVLPEGFLDR 531
Query: 325 ----GMILRGWAPQLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKL 380
G ++ GWAPQ +L HP+ G F++HCG NS LE++ GVPI TWP+ +Q +
Sbjct: 530 TTGIGRVI-GWAPQTRVLTHPATGGFVSHCGWNSTLESIYFGVPIATWPLYAEQQTNAFV 354
Query: 381 MTEVRGVGVEVGAE-EWSMGGGRRERERVVISRDKIEKAVRRLMGGGEEAEQIRRRVHEL 439
+ + VE+ + + GG +++ DKIE +R ++ ++ ++R+RV E+
Sbjct: 353 LVRELKIAVEISLDYRVEINGG----PNYLLTADKIEGGIRSVL---DKDGEVRKRVKEM 195
Query: 440 GNLANAAVQPGGSSYQNLTALI 461
+ + GG SY L LI
Sbjct: 194 SEKSRKTLLEGGCSYSYLDRLI 129
>TC11365 similar to UP|Q8S995 (Q8S995) Glucosyltransferase-14, partial (24%)
Length = 498
Score = 97.8 bits (242), Expect = 3e-21
Identities = 46/105 (43%), Positives = 69/105 (64%)
Frame = +3
Query: 357 EAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRDKIE 416
EA+ GVP++TWP+ GDQF+ EKL+ ++ GVGV VG E + G E V++ ++ +E
Sbjct: 3 EAICGGVPMVTWPLFGDQFFHEKLIVQILGVGVRVGVEA-PVKWGEEEEIGVLVKKEDVE 179
Query: 417 KAVRRLMGGGEEAEQIRRRVHELGNLANAAVQPGGSSYQNLTALI 461
+A+ LM + ++RRRV EL +A AV+ GGSS+ N+T LI
Sbjct: 180 RAIEELMNESSPSHEMRRRVQELAEMAKRAVEEGGSSHTNVTLLI 314
>TC9717 weakly similar to UP|Q9LNI1 (Q9LNI1) F6F3.22 protein, partial (18%)
Length = 902
Score = 80.1 bits (196), Expect(2) = 4e-20
Identities = 47/149 (31%), Positives = 74/149 (49%)
Frame = +2
Query: 294 FVWVVPAEKRGKEEEEEEEKEKWLPEGFEEKGMILRGWAPQLLILGHPSVGAFLTHCGSN 353
F WV+ K+ + + W+ +KGM+ R WAPQ+ IL H S+G FLTHCG +
Sbjct: 98 FFWVLK-----KQNSDSADSHDWIENQSNKKGMVWRTWAPQMRILAHKSIGGFLTHCGWS 262
Query: 354 SALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGAEEWSMGGGRRERERVVISRD 413
S +E + G P+I P +Q + M E + +GV+V + + +RD
Sbjct: 263 SVIEGLQVGCPLIMLPFPNEQMLVARFM-EGKKLGVKVS----------KNQHDGKFTRD 409
Query: 414 KIEKAVRRLMGGGEEAEQIRRRVHELGNL 442
+ KA+R +M EE + R + EL +
Sbjct: 410 SVAKALRSVM-LEEEGKSYRCQAEELSKI 493
Score = 34.7 bits (78), Expect(2) = 4e-20
Identities = 16/32 (50%), Positives = 20/32 (62%)
Frame = +1
Query: 263 SVLYVCFGSMCSFSNEQLHEMACGIEASSVEF 294
SV+YV FGS + SNE E+A G+E S F
Sbjct: 4 SVIYVAFGSEVTLSNEDFTELALGLELSGFPF 99
>AI967812
Length = 274
Score = 93.6 bits (231), Expect = 6e-20
Identities = 45/90 (50%), Positives = 63/90 (70%)
Frame = +2
Query: 334 QLLILGHPSVGAFLTHCGSNSALEAVSAGVPIITWPVLGDQFYTEKLMTEVRGVGVEVGA 393
Q+LIL H ++GAF+THCG NS LE VSAGVP++TWP+ +QFY EKL+T V +GV VG
Sbjct: 17 QVLILEHEAIGAFVTHCGWNSTLEGVSAGVPMVTWPLSAEQFYNEKLVTHVLKIGVPVGV 196
Query: 394 EEWSMGGGRRERERVVISRDKIEKAVRRLM 423
++W+M G I D + K+++R+M
Sbjct: 197 KKWTMFTGDG-----TIKWDAMVKSLKRIM 271
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,243,482
Number of Sequences: 28460
Number of extensions: 144402
Number of successful extensions: 1472
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 1140
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1288
length of query: 481
length of database: 4,897,600
effective HSP length: 94
effective length of query: 387
effective length of database: 2,222,360
effective search space: 860053320
effective search space used: 860053320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0367.1


