

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0359.12
(1065 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
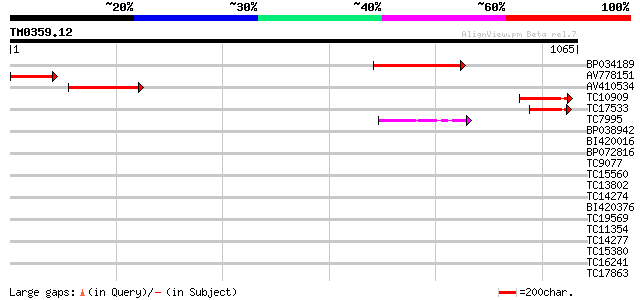
Score E
Sequences producing significant alignments: (bits) Value
BP034189 330 5e-91
AV778151 175 4e-44
AV410534 161 4e-40
TC10909 117 7e-27
TC17533 similar to UP|Q9ZS85 (Q9ZS85) T4B21.1 protein, partial (... 70 2e-12
TC7995 similar to PIR|S63686|S63686 sterol 24-C-methyltransferas... 42 5e-04
BP038942 41 0.001
BI420016 39 0.003
BP072816 39 0.004
TC9077 39 0.004
TC15560 similar to SP|Q03173|NDPP_MOUSE NPC derived proline rich... 38 0.007
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 38 0.010
TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.3... 37 0.013
BI420376 37 0.016
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 37 0.016
TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II ... 37 0.021
TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_1... 37 0.021
TC15380 similar to UP|Q8H5P5 (Q8H5P5) OJ1003_C06.27 protein, par... 36 0.028
TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%) 36 0.037
TC17863 homologue to PIR|S44027|S44027 thioredoxin-disulfide red... 35 0.048
>BP034189
Length = 526
Score = 330 bits (847), Expect = 5e-91
Identities = 173/173 (100%), Positives = 173/173 (100%)
Frame = +1
Query: 684 FETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASP 743
FETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASP
Sbjct: 7 FETAAGKGGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASP 186
Query: 744 PSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLN 803
PSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLN
Sbjct: 187 PSLGRPKDNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLN 366
Query: 804 SLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPS 856
SLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPS
Sbjct: 367 SLVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPS 525
>AV778151
Length = 577
Score = 175 bits (443), Expect = 4e-44
Identities = 88/89 (98%), Positives = 88/89 (98%)
Frame = +2
Query: 1 MESSSRRPFDRSREPGLKRPRMMEEPERVAAAQNPSSRQFLPQRQQQVAASGISTLSSAA 60
MESSSRRPFDRSREPGLKRPRMMEEPERVAAAQNPSSRQFLPQRQQQVAASGISTLSSAA
Sbjct: 311 MESSSRRPFDRSREPGLKRPRMMEEPERVAAAQNPSSRQFLPQRQQQVAASGISTLSSAA 490
Query: 61 RLRAAANSRDLESSDFGRGDGGGGYHPRP 89
RLRAAANSRDLESSDFGRGDGGGG HPRP
Sbjct: 491 RLRAAANSRDLESSDFGRGDGGGGNHPRP 577
>AV410534
Length = 428
Score = 161 bits (408), Expect = 4e-40
Identities = 81/141 (57%), Positives = 102/141 (71%)
Frame = +1
Query: 111 KPIITNLTIIAGENQAAEQAIAATVCANIIEVPSEQKLPSLYLLDSIVKNIGRGYVKYFA 170
KPIIT+LTIIA + + + IA +C I+EV ++QKLPSLYLLDSIVKN+G+ YVKYF+
Sbjct: 4 KPIITDLTIIAEQQREHAEGIADAICTRILEVHADQKLPSLYLLDSIVKNVGQEYVKYFS 183
Query: 171 ARLPEVFCKAYRHVDPAVHHSMRHLFGTWRGVFPSQTLQAIEKELGFTPAVNGSSSTSAT 230
RLPEVFC+AYR V P +H +MRHLFGTW VF + L+ IE +L F+ VN S
Sbjct: 184 LRLPEVFCEAYRQVQPHLHSAMRHLFGTWSKVFSAPVLRKIEAQLQFSRGVNNQPSNVNP 363
Query: 231 LRSDSQSQRPPPSIHVNPKYL 251
LR+ S+S RP IHVNPKY+
Sbjct: 364 LRT-SESPRPTHGIHVNPKYV 423
>TC10909
Length = 569
Score = 117 bits (294), Expect = 7e-27
Identities = 57/100 (57%), Positives = 71/100 (71%)
Frame = +1
Query: 958 KVESLSENEFADSVDRYGKETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFK 1017
K E SENE DSVD + ++TD +Q D MVLADEN CLCV CGELFE+ YCQE WMF+
Sbjct: 7 KSEYQSENESTDSVDVHDEKTDGSQWDTMVLADENPCLCVFCGELFEDVYCQEKDPWMFQ 186
Query: 1018 GAVYVPNSDINDDMGIRDASTGGGPIIHARCLSENPVSSV 1057
G VY+ +SD +M R+ GP++HARCLSENP+S +
Sbjct: 187 GGVYLNHSDSQLEMKSRNV----GPLVHARCLSENPMSHI 294
>TC17533 similar to UP|Q9ZS85 (Q9ZS85) T4B21.1 protein, partial (11%)
Length = 596
Score = 70.1 bits (170), Expect = 2e-12
Identities = 36/79 (45%), Positives = 50/79 (62%)
Frame = +2
Query: 977 ETDRNQEDAMVLADENQCLCVLCGELFEEFYCQENGEWMFKGAVYVPNSDINDDMGIRDA 1036
E ++ E+ V ADE+Q C LCGE F+EFY E EWM++GAVY+ N+ G+ +
Sbjct: 113 EEKKDDEELAVPADEDQNTCALCGEPFDEFYSDETEEWMYRGAVYL-NAPNGTTAGLDRS 289
Query: 1037 STGGGPIIHARCLSENPVS 1055
GPIIHA+C SE+ V+
Sbjct: 290 QL--GPIIHAKCRSESSVA 340
>TC7995 similar to PIR|S63686|S63686 sterol 24-C-methyltransferase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (98%)
Length = 1524
Score = 42.0 bits (97), Expect = 5e-04
Identities = 42/175 (24%), Positives = 73/175 (41%), Gaps = 1/175 (0%)
Frame = +3
Query: 694 LSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVS-AIASPPSLGRPKDN 752
L P+ +P+ S + P+ + S+ + PP+S A SPP R
Sbjct: 123 LHPFPLLEWIPSLSSAPELSSPAAYTGSSAFSAPPNKRANAPPISPADPSPPRKSRTTTT 302
Query: 753 SSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLIS 812
S+ PRR+ +PPR S + S + S + A+ +++P+ + N L +
Sbjct: 303 STGPSSAAPRRS-KPPRRSPISSTPSTISSPTSTSGVGASPSISPLPSPGN------LTA 461
Query: 813 AETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPSTKDEVDDGAKT 867
+T +PS S T SS+ A + G VP P T++ G+++
Sbjct: 462 TPRASTRSMPS------I*SRPSRVTRSSMWDAELVGPCVPSPPTREPTSWGSRS 608
>BP038942
Length = 617
Score = 40.8 bits (94), Expect = 0.001
Identities = 38/112 (33%), Positives = 47/112 (41%)
Frame = +3
Query: 715 PSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALP 774
PS+ PS SP PP +PPS S+ P P + A P S+LP
Sbjct: 177 PSLYSTTPS-----SPPSKQPPPPPTLNPPSCSFSL--STNFPTGPSFKLAYSPTASSLP 335
Query: 775 PASSNLKSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEML 826
P S +LKS + S ++ L ANL SL S T T VPS L
Sbjct: 336 PFSLSLKSGLGTSGSPRHSPLTFSANLSLSL------SPTTNTFTPVPSFFL 473
>BI420016
Length = 559
Score = 39.3 bits (90), Expect = 0.003
Identities = 39/144 (27%), Positives = 62/144 (42%), Gaps = 2/144 (1%)
Frame = +2
Query: 698 PITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALP 757
P T + PT++ GN PS V S S S L PP SPPS P N+ P
Sbjct: 161 PQTLKKPTTQCTHGGN-PSPKPVPASTPSPPSSPTLTPP----PSPPS---PTPNAPLSP 316
Query: 758 KIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPIANLLN--SLVAKGLISAET 815
PP PP + PP+ + + + S++ ++ L P + L+ S + ++
Sbjct: 317 SSPP----SPPTPPSPPPSPAPNPTPSATGSTTPSSPLTPTSASLSSPSSLFSPASTSPA 484
Query: 816 ETTAKVPSEMLTRLEDQSDSITTS 839
TTA + + RL + + +S
Sbjct: 485 STTATTTTTTMIRLRSPASKLFSS 556
>BP072816
Length = 492
Score = 38.9 bits (89), Expect = 0.004
Identities = 33/99 (33%), Positives = 45/99 (45%)
Frame = +1
Query: 694 LSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNS 753
LS+ P + P+S S + LPS + S+ S A P + A PPS P N+
Sbjct: 91 LSHQPCHS--PSSSSSSSSPLPSPPTTLHAPSTSPSSA----PSATAAPPPSTPTPAANT 252
Query: 754 SALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSAN 792
S+ P + P TSA PPASS L + A +N
Sbjct: 253 SSKPS-----TSSKPTTSAAPPASSLLSTPPKPAGPPSN 354
Score = 27.7 bits (60), Expect = 9.9
Identities = 36/150 (24%), Positives = 50/150 (33%), Gaps = 9/150 (6%)
Frame = +1
Query: 641 MTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMPIT 700
++ QP H SS S PLP P TT + S T
Sbjct: 91 LSHQPCHSPSSSSS-------------SSSPLP----SPPTTLHAPSTSPSSAPS---AT 210
Query: 701 NRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIP 760
P S N S +PS SS + + P S +++PP P N S+
Sbjct: 211 AAPPPSTPTPAANTSS----KPSTSSKPTTSAAPPASSLLSTPPKPAGPPSNPSSTNSPA 378
Query: 761 PRRAAQP---------PRTSALPPASSNLK 781
P + P P+ + PA +N K
Sbjct: 379 PASTSAPHAASKAHGSPKVA*TSPACNNSK 468
>TC9077
Length = 745
Score = 38.9 bits (89), Expect = 0.004
Identities = 30/87 (34%), Positives = 45/87 (51%), Gaps = 7/87 (8%)
Frame = +1
Query: 718 LGVRPSQSSGSSPAGLIPPVSAIA---SPPSLGRPKDNSSALPKIPPRR---AAQPPR-T 770
L PS+ S +P+ +PP A A S SL P + S+LPK R PPR T
Sbjct: 232 LSKSPSRPSQHAPSSTLPPPPASAKSSSSTSLHNPLSSPSSLPKPTSRTRRFTPSPPRST 411
Query: 771 SALPPASSNLKSAAVQASSSANNALNP 797
SA P ++S+L++ + SS++ + P
Sbjct: 412 SASPISTSSLENPSSLRSSASKSRSTP 492
>TC15560 similar to SP|Q03173|NDPP_MOUSE NPC derived proline rich protein 1
(NDPP-1). {Mus musculus;}, partial (8%)
Length = 1134
Score = 38.1 bits (87), Expect = 0.007
Identities = 48/191 (25%), Positives = 74/191 (38%), Gaps = 7/191 (3%)
Frame = +3
Query: 695 SNMPITNR------LPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGR 748
S +P TN+ L T+ S + + S PS S +PP A P L
Sbjct: 72 STLPFTNKSHHHYPLTTTASASSSSSSSPSSS*PSPSLPPPSPPPLPPPQAPPLPSHLPS 251
Query: 749 PKDNSSALPKIPPRRAAQP-PRTSALPPASSNLKSAAVQASSSANNALNPIANLLNSLVA 807
P+ +S+ P PP + +P P + P + V SSS + +P + ++ +A
Sbjct: 252 PEPSSTMPPSTPPPQPTKPCPLPKSTPSQPPSSAPHRVPTSSSLASPTSPSSGPHSTTMA 431
Query: 808 KGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSAVPVPSTKDEVDDGAKT 867
S T T PS S T SS AS ++ + E+ D +
Sbjct: 432 APFSSTRTST----PSPNSNNPTPASKPTTFSSLQK*ASTLSFSLRREQSPPEIADPFR- 596
Query: 868 PISLSESTSTE 878
IS S + S+E
Sbjct: 597 -ISSSRNASSE 626
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 37.7 bits (86), Expect = 0.010
Identities = 38/145 (26%), Positives = 65/145 (44%), Gaps = 20/145 (13%)
Frame = +2
Query: 715 PSILGVRPSQ--SSGSSPAGLIP-------PVSAIASP---PSLGRPKDNSSALPKIPPR 762
PS + P++ S+ SSP P P S + +P P+ P + + PK
Sbjct: 86 PSQISSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSPASASPASSPTKSPKPSSP 265
Query: 763 RAAQPPRTSALPPASSNLKSAAVQASSSANNALNP---IANLLNSLVAKGLISAETETTA 819
A+ PP ++ L P+++ +S+ SSS++ + P + L +L K L S+ + ++
Sbjct: 266 SASAPPSSTPLTPSTATSESSPTATSSSSSASPAPPTSFSALSRALELKALASSPSPPSS 445
Query: 820 KVPSE-----MLTRLEDQSDSITTS 839
PS M T L S +TS
Sbjct: 446 PPPSPPSAICMSTSLSSASSVRSTS 520
>TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.35
{Arabidopsis thaliana;} , partial (16%)
Length = 1208
Score = 37.4 bits (85), Expect = 0.013
Identities = 45/158 (28%), Positives = 62/158 (38%), Gaps = 6/158 (3%)
Frame = +3
Query: 686 TAAGKGGKLSNMPITNRLPTSRSLDTGNLPSI-----LGVRPSQSSGSSPAGLIPPVSAI 740
T+ GK S + PT+ S P+ V PS S S P S+
Sbjct: 114 TSTGKPATSSRTSSSGSTPTAVSPPNPTTPATPPPSGTSVSPSLSLTPSLTPFSPSRSST 293
Query: 741 ASPPSLGRPKDNSSALPKIPPRRAAQP-PRTSALPPASSNLKSAAVQASSSANNALNPIA 799
+PP P PP ++ P TS +PPAS+N S+A A+ +A + +
Sbjct: 294 PNPPIPPNPSS--------PPSASSSPTSTTSMIPPASANSPSSAPPAAPTARSTSSSA- 446
Query: 800 NLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSIT 837
SLVA + T T A P+ LT SIT
Sbjct: 447 ----SLVAPN--PSITPTQAPTPTLTLTLTVTLRSSIT 542
Score = 36.2 bits (82), Expect = 0.028
Identities = 46/162 (28%), Positives = 71/162 (43%), Gaps = 12/162 (7%)
Frame = +3
Query: 644 QPSHKQQ------LGSSHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNM 697
+P K+Q L S T+ + P +S+ T +P T+ +++G +
Sbjct: 18 EPRRKEQ*HHLVLLRRSRWISTSPSSPPSISRT----STGKPATSSRTSSSGSTPTAVSP 185
Query: 698 P--ITNRLP----TSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKD 751
P T P TS S PS+ PS+SS +P IPP +SPPS
Sbjct: 186 PNPTTPATPPPSGTSVSPSLSLTPSLTPFSPSRSS--TPNPPIPPNP--SSPPSASSSPT 353
Query: 752 NSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANN 793
+++++ IPP A P S+ PPA+ +S + AS A N
Sbjct: 354 STTSM--IPPASANSP---SSAPPAAPTARSTSSSASLVAPN 464
>BI420376
Length = 562
Score = 37.0 bits (84), Expect = 0.016
Identities = 29/78 (37%), Positives = 38/78 (48%), Gaps = 3/78 (3%)
Frame = +2
Query: 704 PTSRSLDTGNLPSILGVRP---SQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIP 760
PTS SL + LPS P S +SG++ PP SA SPPS +++P P
Sbjct: 77 PTSLSLGSTTLPSPSFSAPSSTSSASGNTQQTPPPPPSAATSPPSTA-----PTSIPPPP 241
Query: 761 PRRAAQPPRTSALPPASS 778
P PP S PPA++
Sbjct: 242 P----LPPTPSISPPATT 283
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 37.0 bits (84), Expect = 0.016
Identities = 41/157 (26%), Positives = 58/157 (36%), Gaps = 2/157 (1%)
Frame = +1
Query: 633 LQSGSFSSMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGG 692
L + S SS ++ P +H+ T+ T P + TE P T +
Sbjct: 37 LHTSSSSSSSYSPPPPPSQQPTHSPSTSPTSTTTPPPSPT-KVTENPPTAPSTSTKSPTS 213
Query: 693 KLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPS--LGRPK 750
S P P + S T P P S SP PP +A+ASP + L K
Sbjct: 214 SASEEPS----PPNPSTSTTQQPKHSPTSPPASPSPSPPSTKPP-TAMASPSTSPLSATK 378
Query: 751 DNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQA 787
+ + P P + PP T++L SS S A
Sbjct: 379 SHPTP-PAAPSASSTPPPTTTSLKTMSSPSSSTPSSA 486
>TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II , partial
(6%)
Length = 583
Score = 36.6 bits (83), Expect = 0.021
Identities = 40/146 (27%), Positives = 64/146 (43%), Gaps = 6/146 (4%)
Frame = -1
Query: 738 SAIASPPSLGRPKDNSSALPKIPPRRAAQP-----PRTSALPPASSNLKSAAVQASSSAN 792
S+ A P S GR K + +A I P A++P P + PP S + + + SS
Sbjct: 445 SSSAPPSSPGRKKSSETA---IRPHSASEPDPAPPPPPPSSPPMSPSPTATSPPTSSPNT 275
Query: 793 NALNPIANLLNS-LVAKGLISAETETTAKVPSEMLTRLEDQSDSITTSSSLPGASVSGSA 851
+ P N +S + S+ + +T +PS TR S+S T+SS SGS+
Sbjct: 274 HPPTPTPNP*SSPSPSTTTASSASPSTKTLPS---TRPRPASESPTSSSPSSPPPSSGSS 104
Query: 852 VPVPSTKDEVDDGAKTPISLSESTST 877
V P + +P + S++T
Sbjct: 103 VSPPRISPPPPPPSNSPTATPPSSAT 26
Score = 35.8 bits (81), Expect = 0.037
Identities = 34/131 (25%), Positives = 50/131 (37%), Gaps = 3/131 (2%)
Frame = -1
Query: 645 PSHKQQLGSSHTEVTAKTEKPHL---SKVPLPRETEQPTTTRFETAAGKGGKLSNMPITN 701
P + S T + T P+ + P P + P+TT +A+ P T
Sbjct: 343 PPSSPPMSPSPTATSPPTSSPNTHPPTPTPNP*SSPSPSTTTASSAS---------PSTK 191
Query: 702 RLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPP 761
LP++R P P+ SS SSP P + SPP + P S P P
Sbjct: 190 TLPSTR-------PRPASESPTSSSPSSPP---PSSGSSVSPPRISPPPPPPSNSPTATP 41
Query: 762 RRAAQPPRTSA 772
+A P +S+
Sbjct: 40 PSSATNPSSSS 8
Score = 32.3 bits (72), Expect = 0.40
Identities = 36/155 (23%), Positives = 58/155 (37%), Gaps = 3/155 (1%)
Frame = -1
Query: 639 SSMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGKGGKLSNMP 698
SS + PS + SS T + +PH + P P P ++ +S P
Sbjct: 448 SSSSAPPSSPGRKKSSETAI-----RPHSASEPDPAPPPPPPSSP---------PMSPSP 311
Query: 699 ITNRLPTSRSLDTGNLPSI---LGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSA 755
PTS P+ PS ++ SS + + + P+ P +S +
Sbjct: 310 TATSPPTSSPNTHPPTPTPNP*SSPSPSTTTASSASPSTKTLPSTRPRPASESPTSSSPS 131
Query: 756 LPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSS 790
P + PPR S PP SN +A +S++
Sbjct: 130 SPPPSSGSSVSPPRISPPPPPPSNSPTATPPSSAT 26
>TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_13), partial
(33%)
Length = 941
Score = 36.6 bits (83), Expect = 0.021
Identities = 40/168 (23%), Positives = 64/168 (37%)
Frame = +3
Query: 631 RDLQSGSFSSMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRETEQPTTTRFETAAGK 690
R S S S P H SS + T P S P PR P +T
Sbjct: 93 RSPPSSSSSPSPPPPPHAPAAASSSPPTPSATLPPTPSP-PSPRSDPSPLSTS------- 248
Query: 691 GGKLSNMPITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPK 750
+ P T+ T ++ + P+ + + GS PP+ SPPS+ P+
Sbjct: 249 ----TTNPTTSSSTTPKTTTNKSNPNTARLSHAAP*GS------PPLLTTTSPPSVTAPR 398
Query: 751 DNSSALPKIPPRRAAQPPRTSALPPASSNLKSAAVQASSSANNALNPI 798
+S++ ++P A + LP SS S V ++ A++P+
Sbjct: 399 TSSAS--RLPSSSALDAALSPPLPCTSSGPSSPPVTTTAPPCTAISPM 536
>TC15380 similar to UP|Q8H5P5 (Q8H5P5) OJ1003_C06.27 protein, partial (28%)
Length = 1306
Score = 36.2 bits (82), Expect = 0.028
Identities = 44/164 (26%), Positives = 67/164 (40%), Gaps = 8/164 (4%)
Frame = +2
Query: 722 PSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSA-LPPASSNL 780
PS SS SP P++A +P + +++ P+ P A PP TS PPA+S
Sbjct: 122 PSSSSPPSPT----PLAAPTAPSTAASSTSSTARFPQTPNPSTASPPPTSPWTPPATSGS 289
Query: 781 KSAAVQASSSANNALNPIANLLNSLVAKGLISAETETTAKVPSEMLTRLEDQSDSI---- 836
S P +L +S TA+V L R + + S
Sbjct: 290 ASLPPPPPPPP----RPFPSLCSS-------------TAEVSPSCLRRRSNSTPSAASSA 418
Query: 837 --TTSSSLPGASVS-GSAVPVPSTKDEVDDGAKTPISLSESTST 877
+T SS P +VS S V +P+T+ ++ P+S T+T
Sbjct: 419 VPSTPSSSPSTTVSPPSTVTLPNTRTDL-----MPLSSLTKTAT 535
>TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%)
Length = 799
Score = 35.8 bits (81), Expect = 0.037
Identities = 44/158 (27%), Positives = 67/158 (41%), Gaps = 6/158 (3%)
Frame = +1
Query: 640 SMTFQPSHKQQLGSSHTEVTAKTEKPHLSKVPLPRE-TEQPTTTRFETAAGKGGKLSNMP 698
S T QP + Q ++ + ++ P+LS P T QP + A S+ P
Sbjct: 94 SPTSQPPNTQTPANTPSP---SSQPPYLSTPPKSNSPTSQPPVAQTPRAVTPISSPSSSP 264
Query: 699 ITNRLPTSRSLDTGNLPSILGVRPSQSSGSSPAGLIP---PVSAIASPPSLGRPKDNSSA 755
+N S N PS+ + S +SP L P PV A ASPPS ++ A
Sbjct: 265 TSNSPYPST-----NPPSL-----APSISTSPPALTPASTPVPATASPPSQSPSAESPGA 414
Query: 756 -LPKIP-PRRAAQPPRTSALPPASSNLKSAAVQASSSA 791
LP P ++ PR++ P +++ A +SSA
Sbjct: 415 SLPAATTPSPSSTTPRSTQTPSSNNETTPAQRNGASSA 528
>TC17863 homologue to PIR|S44027|S44027 thioredoxin-disulfide reductase 2
[validated] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (19%)
Length = 334
Score = 35.4 bits (80), Expect = 0.048
Identities = 23/79 (29%), Positives = 40/79 (50%)
Frame = +3
Query: 715 PSILGVRPSQSSGSSPAGLIPPVSAIASPPSLGRPKDNSSALPKIPPRRAAQPPRTSALP 774
P L R S+ S +S + PP ++PP+ P N S+ P PP ++ P
Sbjct: 105 PPWLTTRSSKPSSASSEAVPPPTPPPSTPPA---PSSNQSSSKAGWP--TTSPPAANSPP 269
Query: 775 PASSNLKSAAVQASSSANN 793
P +S + A+++AS +A++
Sbjct: 270 PPTSRISPASLKASWAASS 326
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,734,225
Number of Sequences: 28460
Number of extensions: 263707
Number of successful extensions: 2392
Number of sequences better than 10.0: 237
Number of HSP's better than 10.0 without gapping: 2073
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2272
length of query: 1065
length of database: 4,897,600
effective HSP length: 100
effective length of query: 965
effective length of database: 2,051,600
effective search space: 1979794000
effective search space used: 1979794000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0359.12


