

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
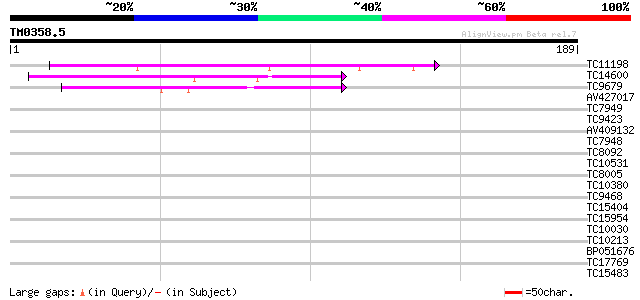
Query= TM0358.5
(189 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11198 similar to UP|Q9LJT8 (Q9LJT8) Non-race specific disease ... 52 5e-08
TC14600 similar to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protei... 40 2e-04
TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, p... 40 3e-04
AV427017 34 0.018
TC7949 33 0.024
TC9423 similar to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein... 32 0.053
AV409132 32 0.090
TC7948 32 0.090
TC8092 similar to UP|Q8MLI6 (Q8MLI6) CG30127-PA, partial (3%) 29 0.45
TC10531 similar to UP|Q96UV7 (Q96UV7) Endo-1,4-beta-xylanase , ... 29 0.58
TC8005 similar to UP|Q9XQB2 (Q9XQB2) Chlorophyll a/b binding pro... 28 0.76
TC10380 similar to UP|Q8S8U2 (Q8S8U2) Ycf1 protein, partial (40%) 28 1.3
TC9468 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (72%) 27 1.7
TC15404 similar to UP|GSA_SOYBN (P45621) Glutamate-1-semialdehyd... 27 1.7
TC15954 similar to UP|Q9LJE5 (Q9LJE5) Gb|AAD10646.1 (AT3g13460/M... 26 3.8
TC10030 similar to UP|Q94JM3 (Q94JM3) AT5g62000/mtg10_20, partia... 26 3.8
TC10213 homologue to UP|PFPB_RICCO (Q41141) Pyrophosphate--fruct... 26 4.9
BP051676 25 6.4
TC17769 similar to UP|Q40175 (Q40175) Transcription factor, part... 25 6.4
TC15483 homologue to UP|AAR17698 (AAR17698) GTP-binding protein ... 25 8.4
>TC11198 similar to UP|Q9LJT8 (Q9LJT8) Non-race specific disease resistance
protein-like, partial (17%)
Length = 882
Score = 52.4 bits (124), Expect = 5e-08
Identities = 39/147 (26%), Positives = 67/147 (45%), Gaps = 17/147 (11%)
Frame = +2
Query: 14 FIGFLGLLVLCLWLALRPKSPSYSVVFL-------SIEQHPGENGSIFYSLEIENPNKDS 66
FI +GL L +WL+LR P + ++ ++ N ++ ++L++ NPNKD
Sbjct: 98 FITTMGLAALFIWLSLRVDEPKCYIDYIYIPALNKTLNSPFTTNSTLNFTLKLVNPNKDK 277
Query: 67 SIYYDDIILSFLYGQQEDK---VGETTIGSFHQGTGKTRSVYDTVNARHGAF------KP 117
I YD + L F + +G T+G F+QG K ++NA G K
Sbjct: 278 GIKYDAVRLRFAIFVDHNTTRPIGNATVGGFYQGHEKKARKPGSLNAAAGNLTAKVDGKM 457
Query: 118 LFKAISNATAELKVAL-TTRFQYKTWG 143
F+ + + K++L T+ + WG
Sbjct: 458 YFRVEFDTAVKYKISLWYTKKRDLLWG 538
>TC14600 similar to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein B13-1-9,
partial (76%)
Length = 1052
Score = 40.4 bits (93), Expect = 2e-04
Identities = 30/114 (26%), Positives = 54/114 (47%), Gaps = 8/114 (7%)
Frame = +1
Query: 7 FYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHPGENGSIFYSLEIE----NP 62
F+ L+ I +GL++L +L ++P+ + V + + N S+ Y++ + NP
Sbjct: 244 FWKILITLIVLVGLVILIFYLIVQPRPFKFYVNEAKLTEFNYTNNSLRYNMVLNFTARNP 423
Query: 63 NKDSSIYYDDIILSFLYGQ----QEDKVGETTIGSFHQGTGKTRSVYDTVNARH 112
NK +IYYD + Y +DKV T + SF Q + + T + +H
Sbjct: 424 NKKLNIYYDKVEALAFYEDSRFASDDKV-ITHMNSFRQYKKSSSPMSATFSGQH 582
>TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partial
(54%)
Length = 710
Score = 40.0 bits (92), Expect = 3e-04
Identities = 29/100 (29%), Positives = 43/100 (43%), Gaps = 5/100 (5%)
Frame = +2
Query: 18 LGLLVLCLWLALRPKSPSYSVVFLSIEQHPGE-NGSIFYSLE----IENPNKDSSIYYDD 72
+G+ VL WL +RP + V ++ Q N ++ Y L + NPNK IYYD
Sbjct: 218 IGIAVLIFWLIVRPNVVKFHVTDATLTQFNFTGNNTLNYDLTLNITVRNPNKRLGIYYDY 397
Query: 73 IILSFLYGQQEDKVGETTIGSFHQGTGKTRSVYDTVNARH 112
I +Y Q+ + F+QG KT + H
Sbjct: 398 IEARAMY--QDVRFDTEFFDRFYQGHKKTNVISRQFKGTH 511
>AV427017
Length = 429
Score = 33.9 bits (76), Expect = 0.018
Identities = 17/58 (29%), Positives = 27/58 (46%), Gaps = 4/58 (6%)
Frame = +2
Query: 26 WLALRPKSPSYSVVFLSIEQHPGENGSIFYSLE----IENPNKDSSIYYDDIILSFLY 79
WL +RP + V S+ + N ++ Y+L + NPN +YYD I + Y
Sbjct: 221 WLIVRPNVVKFRVTEASLTEFTYTNNTLHYNLALNITVRNPNSRVGLYYDSIETTAFY 394
>TC7949
Length = 1183
Score = 33.5 bits (75), Expect = 0.024
Identities = 21/98 (21%), Positives = 43/98 (43%), Gaps = 5/98 (5%)
Frame = +1
Query: 5 KRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHPGENGSIFYS-----LEI 59
+R + ++ F+ + + +L +W L+P P++++ +++ + S L
Sbjct: 250 RRIFWGIVIFLFIVLVTILIIWAILKPNKPTFTLQDVTVYAFNATMANFLTSNFQVTLSA 429
Query: 60 ENPNKDSSIYYDDIILSFLYGQQEDKVGETTIGSFHQG 97
NPN +YYD + Y Q + T I +QG
Sbjct: 430 RNPNDKIGVYYDRLDAYITY-QSQQVTYRTAIPPSYQG 540
>TC9423 similar to UP|Q8S8Z8 (Q8S8Z8) Syringolide-induced protein B13-1-9,
partial (53%)
Length = 969
Score = 32.3 bits (72), Expect = 0.053
Identities = 21/75 (28%), Positives = 37/75 (49%), Gaps = 12/75 (16%)
Frame = +1
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQ--------HPGENGSIFYSLEIE-- 60
L+ I FL L+ L +L ++P++ + V + Q + N + Y+L +
Sbjct: 190 LLAIILFLALVFLIFYLIVQPRAFKFHVTDAKLTQFNYTTTNNNNSNNHLLRYNLVLNFT 369
Query: 61 --NPNKDSSIYYDDI 73
NPNK +IYYD++
Sbjct: 370 ARNPNKKLNIYYDEV 414
>AV409132
Length = 405
Score = 31.6 bits (70), Expect = 0.090
Identities = 23/82 (28%), Positives = 38/82 (46%), Gaps = 16/82 (19%)
Frame = +1
Query: 5 KRFYVFLMEFIGFLGLLVLCLWLALRPKSPSY----SVVF---LSIEQH------PGENG 51
+R ++ FI + L++ +W+ LRP P + + VF LS +QH P N
Sbjct: 139 RRILAAVVAFIFLILLIIFLIWIILRPTKPRFMLQDATVFAFNLSTQQHTPPLLTPTPNT 318
Query: 52 ---SIFYSLEIENPNKDSSIYY 70
++ +L NPN +YY
Sbjct: 319 LTLTLQVTLSSHNPNARIGVYY 384
>TC7948
Length = 1230
Score = 31.6 bits (70), Expect = 0.090
Identities = 17/84 (20%), Positives = 38/84 (45%), Gaps = 5/84 (5%)
Frame = +2
Query: 5 KRFYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHPGENGSIF-----YSLEI 59
+R + ++ F+ + + +L +W LRP P++ + +++ + +L
Sbjct: 395 RRVFCCIVVFLFIVLVTILLIWAILRPTKPTFILQDVTVYAFNATVPNFLTTNFQVTLTS 574
Query: 60 ENPNKDSSIYYDDIILSFLYGQQE 83
NPN+ +YYD + Y Q+
Sbjct: 575 RNPNEHIGVYYDRLSTYVTYRSQQ 646
>TC8092 similar to UP|Q8MLI6 (Q8MLI6) CG30127-PA, partial (3%)
Length = 1073
Score = 29.3 bits (64), Expect = 0.45
Identities = 20/70 (28%), Positives = 32/70 (45%), Gaps = 10/70 (14%)
Frame = +3
Query: 11 LMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIE----QHPGENGSIFYSLEIE------ 60
L F+ LG+ +L RPK+P+YS+ +S++ P + + S E +
Sbjct: 321 LFIFLILLGIAAGVFYLVFRPKAPNYSIQTVSVKGMNLTSPSPSSAAAISPEFDVAVRAN 500
Query: 61 NPNKDSSIYY 70
N N IYY
Sbjct: 501 NGNDKIGIYY 530
>TC10531 similar to UP|Q96UV7 (Q96UV7) Endo-1,4-beta-xylanase , partial
(6%)
Length = 387
Score = 28.9 bits (63), Expect = 0.58
Identities = 16/72 (22%), Positives = 37/72 (51%), Gaps = 6/72 (8%)
Frame = +2
Query: 5 KRFYVFLMEFIGFLGLLVLCLWLALRPKSPSY--SVVFLSIEQHPGENGSIFYSL----E 58
+ F++ L + L++L +W+ ++P+S ++ S L+ + ++ ++L
Sbjct: 170 RTFWILLATILLLACLIILVVWILIQPRSYTFHASQAKLTEFNYNSTTATLRHNLVVNYT 349
Query: 59 IENPNKDSSIYY 70
+ NPNK IY+
Sbjct: 350 VRNPNKKLKIYF 385
>TC8005 similar to UP|Q9XQB2 (Q9XQB2) Chlorophyll a/b binding protein CP29,
complete
Length = 1116
Score = 28.5 bits (62), Expect = 0.76
Identities = 30/125 (24%), Positives = 52/125 (41%), Gaps = 17/125 (13%)
Frame = -1
Query: 82 QEDKVGETTIGSFHQGTG----KTRSVYDTVNARHGAFKPLFKAISNATAELKVALTTRF 137
+E G+T++ F Q G + R + V +PL + + + EL V F
Sbjct: 816 KESNHGKTSMLDFCQLKGGLLLRVRGQAERVEEAATWVQPLLRVKLSVSLELNVPNHQNF 637
Query: 138 QYKTWG---------IKSKFHGLHLQGILPIDS----DGKLSRKKKKYPLSRSSRKLARS 184
G + F+ LHL G+LP DS +GK +++ + P S + L +
Sbjct: 636 NPNQCGD*EWEW*TKV**PFYKLHLAGVLPGDSGEPLNGKSTKRGEHGPPSMNKLALPKP 457
Query: 185 IAKRH 189
+ +H
Sbjct: 456 LQPKH 442
>TC10380 similar to UP|Q8S8U2 (Q8S8U2) Ycf1 protein, partial (40%)
Length = 5664
Score = 27.7 bits (60), Expect = 1.3
Identities = 17/40 (42%), Positives = 23/40 (57%)
Frame = +2
Query: 133 LTTRFQYKTWGIKSKFHGLHLQGILPIDSDGKLSRKKKKY 172
L T+F K+ F GL+L P D+ GK+S KKKK+
Sbjct: 1601 LITKFSRKSVS-SLNFEGLYL---FPKDNKGKMSSKKKKF 1708
>TC9468 similar to UP|Q9FMN3 (Q9FMN3) Emb|CAB53482.1, partial (72%)
Length = 1317
Score = 27.3 bits (59), Expect = 1.7
Identities = 14/46 (30%), Positives = 21/46 (45%)
Frame = +3
Query: 7 FYVFLMEFIGFLGLLVLCLWLALRPKSPSYSVVFLSIEQHPGENGS 52
F +FL+ F+ L L LW A RP P + + + + GS
Sbjct: 387 FLIFLLGFLLLFTLFSLILWGASRPMKPKIFIKSIKFDHVQVQAGS 524
>TC15404 similar to UP|GSA_SOYBN (P45621) Glutamate-1-semialdehyde
2,1-aminomutase, chloroplast precursor (GSA)
(Glutamate-1-semialdehyde aminotransferase) (GSA-AT) ,
partial (39%)
Length = 801
Score = 27.3 bits (59), Expect = 1.7
Identities = 14/36 (38%), Positives = 21/36 (57%), Gaps = 2/36 (5%)
Frame = +1
Query: 5 KRFYVFLMEFIGFLGLLVLCLW--LALRPKSPSYSV 38
+ F+ F+ F+G L++L LW L L P S S+ V
Sbjct: 592 REFFYFIFSFVGQFRLILLLLW*FLLL*PVSLSHDV 699
>TC15954 similar to UP|Q9LJE5 (Q9LJE5) Gb|AAD10646.1 (AT3g13460/MRP15_10),
partial (7%)
Length = 770
Score = 26.2 bits (56), Expect = 3.8
Identities = 10/24 (41%), Positives = 15/24 (61%)
Frame = +3
Query: 138 QYKTWGIKSKFHGLHLQGILPIDS 161
Q++ WG+ + GL+ ILPI S
Sbjct: 666 QFQPWGMMVSYMGLNTTSILPISS 737
>TC10030 similar to UP|Q94JM3 (Q94JM3) AT5g62000/mtg10_20, partial (13%)
Length = 588
Score = 26.2 bits (56), Expect = 3.8
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = +1
Query: 12 MEFIGFLGLLVLCLWLALRPKSPSYSV 38
+ F G G+LVL LWL + K + S+
Sbjct: 340 LHFTGSYGMLVLVLWLLCQEKESASSI 420
>TC10213 homologue to UP|PFPB_RICCO (Q41141) Pyrophosphate--fructose
6-phosphate 1-phosphotransferase beta subunit (PFP)
(6-phosphofructokinase, pyrophosphate-dependent)
(Pyrophosphate-dependent 6-phosphofructose-1-kinase)
(PPi-PFK) , partial (22%)
Length = 572
Score = 25.8 bits (55), Expect = 4.9
Identities = 24/65 (36%), Positives = 32/65 (48%), Gaps = 2/65 (3%)
Frame = +3
Query: 86 VGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAISNATAELKVALTTRFQ--YKTWG 143
V E T+G GT T S+ D V RHG FKP+ K A EL A +F W
Sbjct: 132 VEEWTVG----GTALT-SLMD-VERRHGKFKPVIK---KAMVELDGAPFKKFASLRDEWA 284
Query: 144 IKSKF 148
+K+++
Sbjct: 285 LKNRY 299
>BP051676
Length = 564
Score = 25.4 bits (54), Expect = 6.4
Identities = 24/63 (38%), Positives = 32/63 (50%), Gaps = 2/63 (3%)
Frame = -2
Query: 86 VGETTIGSFHQGTGKTRSVYDTVNARHGAFKPLFKAISNATAELKVALTTRFQ--YKTWG 143
V E T+G GT T S+ D V RHG FKP+ K A EL+ A +F + W
Sbjct: 455 VKEWTVG----GTALT-SLMD-VERRHGKFKPVIK---KAMVELEGAPFKKFASLREEWA 303
Query: 144 IKS 146
+K+
Sbjct: 302 LKN 294
>TC17769 similar to UP|Q40175 (Q40175) Transcription factor, partial (8%)
Length = 545
Score = 25.4 bits (54), Expect = 6.4
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = -3
Query: 40 FLSIEQHPGENGSIFYSLEIENPNKDSSI 68
FLS ++ GEN S F + PNKDS++
Sbjct: 204 FLSGDKLDGEN*SEFGDSKDSKPNKDSNV 118
>TC15483 homologue to UP|AAR17698 (AAR17698) GTP-binding protein TypA,
partial (11%)
Length = 615
Score = 25.0 bits (53), Expect = 8.4
Identities = 9/18 (50%), Positives = 14/18 (77%)
Frame = -2
Query: 21 LVLCLWLALRPKSPSYSV 38
+V+C ++ L KSPSY+V
Sbjct: 326 VVICFFITLAAKSPSYAV 273
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,259,766
Number of Sequences: 28460
Number of extensions: 42363
Number of successful extensions: 221
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 219
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 219
length of query: 189
length of database: 4,897,600
effective HSP length: 85
effective length of query: 104
effective length of database: 2,478,500
effective search space: 257764000
effective search space used: 257764000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0358.5


