

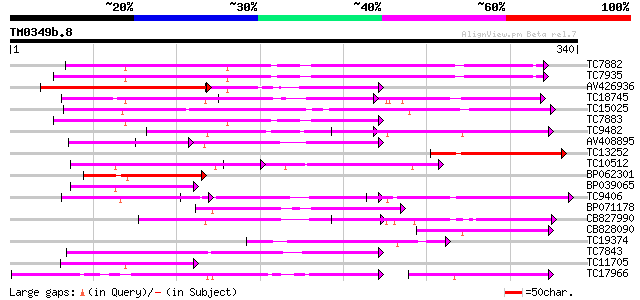
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0349b.8
(340 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC7882 homologue to UP|O49875 (O49875) Adenine nucleotide transl... 129 9e-31
TC7935 similar to UP|O49447 (O49447) ADP, ATP carrier-like prote... 125 1e-29
AV426936 121 1e-28
TC18745 weakly similar to UP|ADT2_HUMAN (P05141) ADP,ATP carrier... 115 1e-26
TC15025 similar to UP|MCAT_ARATH (Q93XM7) Mitochondrial carnitin... 97 3e-21
TC7883 homologue to UP|ADT1_GOSHI (O22342) ADP,ATP carrier prote... 96 7e-21
TC9482 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier prote... 92 1e-19
AV408895 84 4e-17
TC13252 similar to UP|Q9MA27 (Q9MA27) T5E21.6, partial (17%) 84 4e-17
TC10512 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier prot... 65 2e-11
BP062301 64 4e-11
BP039065 64 4e-11
TC9406 similar to UP|Q9M058 (Q9M058) Ca-dependent solute carrier... 64 5e-11
BP071178 64 5e-11
CB827990 63 8e-11
CB828090 62 2e-10
TC19374 similar to UP|Q8LNZ1 (Q8LNZ1) Mitochondrial uncoupling p... 56 1e-08
TC7843 weakly similar to GB|AAF03412.1|6090963|AF188712 mitochon... 55 2e-08
TC11705 homologue to UP|O49875 (O49875) Adenine nucleotide trans... 52 1e-07
TC17966 similar to UP|Q8L9F8 (Q8L9F8) Mitochondrial carrier-like... 50 5e-07
>TC7882 homologue to UP|O49875 (O49875) Adenine nucleotide translocator,
complete
Length = 1656
Score = 129 bits (323), Expect = 9e-31
Identities = 94/300 (31%), Positives = 145/300 (48%), Gaps = 10/300 (3%)
Frame = +1
Query: 34 ELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-------LGVCQSLNKLVKHEGFLG 86
+ + GGV+ A+SKTA AP+ERVK+L Q + T G+ + K EG +
Sbjct: 463 DFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKTGRLSEPYKGIGDCFARTTKDEGIVA 642
Query: 87 LYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFI--DLLAGSAAGGTSVLC 144
L++GN A+ +R P AL+F + +K G + +L +G AAG +S+L
Sbjct: 643 LWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDRDGYWKWFAGNLASGGAAGASSLLF 822
Query: 145 TYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGP 204
Y LD ART+LA D + K G + G+ V G GLYRG
Sbjct: 823 VYSLDYARTRLAN---DAKAAKKGGERQFN------GLVDVYRKTLASDGVAGLYRGFNI 975
Query: 205 TITGILPYAGLKFYTYEKLK-MHVPEEHQKSILMRLSCGALAGLFGQTLTYPLDVVKRQM 263
+ GI+ Y GL F Y+ LK + + + Q S G L +YP+D V+R+M
Sbjct: 976 SCVGIIVYRGLYFGMYDSLKPVLLTGKLQDSFFASFGLGWLITNGAGLASYPIDTVRRRM 1155
Query: 264 QVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMK 323
+ S E +Y+++LD ++I++N+G + LF G N +R V A + YD ++
Sbjct: 1156MMTS-----GEAVKYKSSLDAFQQILKNEGAKSLFKGAGANILRAVAGAGV-LAGYDKLQ 1317
>TC7935 similar to UP|O49447 (O49447) ADP, ATP carrier-like protein,
partial (83%)
Length = 1465
Score = 125 bits (313), Expect = 1e-29
Identities = 91/307 (29%), Positives = 147/307 (47%), Gaps = 10/307 (3%)
Frame = +3
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-------LGVCQSLNKLV 79
G ++ + + GGV+ A+SKTA AP+ER+K+L Q + + G+ +
Sbjct: 231 GASAFLVDFLMGGVSAAVSKTAAAPIERIKLLIQNQDEMIKSGRLSEPYKGIGDCFARTT 410
Query: 80 KHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFI--DLLAGSAA 137
K EG + L++GN A+ +R P AL+F + +K G + +L +G AA
Sbjct: 411 KDEGAIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAA 590
Query: 138 GGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARG 197
G +S+L Y LD ART+LA D++ K G + G+ V + G G
Sbjct: 591 GASSLLFVYSLDYARTRLAN---DSKSAKKGGERQFN------GLVDVYKKTIQSDGIAG 743
Query: 198 LYRGVGPTITGILPYAGLKFYTYEKLK-MHVPEEHQKSILMRLSCGALAGLFGQTLTYPL 256
LYRG + GI+ Y GL F Y+ LK + + + Q S G + +YP+
Sbjct: 744 LYRGFSISCIGIIVYRGLYFGMYDSLKPVVLVGDMQDSFFASFLLGWGITIGAGLASYPI 923
Query: 257 DVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISF 316
D V+R+M + S E +Y+++LD + I+ +G + LF G N +R V A +
Sbjct: 924 DTVRRRMMMTS-----GEAVKYKSSLDAFKVIIAKEGTKSLFKGAGANILRAVAGAGV-L 1085
Query: 317 TAYDTMK 323
YD ++
Sbjct: 1086AGYDKLQ 1106
>AV426936
Length = 401
Score = 121 bits (304), Expect = 1e-28
Identities = 57/103 (55%), Positives = 73/103 (70%)
Frame = +2
Query: 19 QKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKL 78
Q + D VPV+ KEL+AGG+AG +K+ VAPLERVKIL QTR F + G+ S ++
Sbjct: 80 QNNAVLLDHVPVFAKELLAGGIAGGFAKSVVAPLERVKILLQTRRAEFQSSGLWGSATRI 259
Query: 79 VKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCP 121
K EGFLG Y+GNGAS RI+PYAALH+M+YE Y+ WI+ P
Sbjct: 260 AKTEGFLGFYRGNGASVARIIPYAALHYMSYEEYRRWIVLTFP 388
Score = 41.6 bits (96), Expect = 2e-04
Identities = 34/109 (31%), Positives = 49/109 (44%), Gaps = 2/109 (1%)
Frame = +2
Query: 118 NNCPALGSGPFI--DLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRP 175
NN L P +LLAG AGG + PL+ R K+ Q TR R
Sbjct: 83 NNAVLLDHVPVFAKELLAGGIAGGFAKSVVAPLE--RVKILLQ---TR----------RA 217
Query: 176 QPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLK 224
+ G+ T + + G G YRG G ++ I+PYA L + +YE+ +
Sbjct: 218 EFQSSGLWGSATRIAKTEGFLGFYRGNGASVARIIPYAALHYMSYEEYR 364
>TC18745 weakly similar to UP|ADT2_HUMAN (P05141) ADP,ATP carrier protein,
fibroblast isoform (ADP/ATP translocase 2) (Adenine
nucleotide translocator 2) (ANT 2), partial (9%)
Length = 834
Score = 115 bits (287), Expect = 1e-26
Identities = 74/202 (36%), Positives = 106/202 (51%), Gaps = 12/202 (5%)
Frame = +3
Query: 32 VKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-------LGVCQSLNKLVKHEGF 84
V +L+AGGVAGA SKT APL R+ IL+Q + G H+ + Q ++++ EGF
Sbjct: 276 VNQLLAGGVAGAFSKTCTAPLARLTILFQVQ--GMHSDFSALRKPSIWQEASRIINEEGF 449
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWI-----LNNCPALGSGPFIDLLAGSAAGG 139
KGN + V +PY+A+ F +YERYK+ N + + F+ + G AG
Sbjct: 450 RAFGKGNMVTIVHRLPYSAVSFYSYERYKNLFHSLMGENYRGSSSANLFVHFMGGGLAGV 629
Query: 140 TSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLY 199
TS TYPLDL RT+LA Q R TI + GI +++ R+ G GLY
Sbjct: 630 TSASATYPLDLVRTRLAAQ----RSTI-----------YYRGISHAFSTICRDEGFLGLY 764
Query: 200 RGVGPTITGILPYAGLKFYTYE 221
+G+G T+ G+ P + F YE
Sbjct: 765 KGLGATLLGVGPSIAISFSVYE 830
Score = 73.9 bits (180), Expect = 4e-14
Identities = 62/204 (30%), Positives = 94/204 (45%), Gaps = 8/204 (3%)
Frame = +3
Query: 126 GPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSV 185
G LLAG AG S CT PL AR + +QV +G D +P I
Sbjct: 270 GTVNQLLAGGVAGAFSKTCTAPL--ARLTILFQV---QGMHSDFSALRKPS-----IWQE 419
Query: 186 LTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLK--MH--VPEEHQKS----ILM 237
+ + E G R +G TI LPY+ + FY+YE+ K H + E ++ S + +
Sbjct: 420 ASRIINEEGFRAFGKGNMVTIVHRLPYSAVSFYSYERYKNLFHSLMGENYRGSSSANLFV 599
Query: 238 RLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQL 297
G LAG+ + TYPLD+V+ ++ A YR I R++G+ L
Sbjct: 600 HFMGGGLAGVTSASATYPLDLVRTRLA------AQRSTIYYRGISHAFSTICRDEGFLGL 761
Query: 298 FAGVSINYIRIVPSAAISFTAYDT 321
+ G+ + + PS AISF+ Y++
Sbjct: 762 YKGLGATLLGVGPSIAISFSVYES 833
>TC15025 similar to UP|MCAT_ARATH (Q93XM7) Mitochondrial
carnitine/acylcarnitine carrier-like protein (A BOUT DE
SOUFFLE) (Carnitine/acylcarnitine translocase-like
protein) (CAC-like protein), partial (97%)
Length = 1256
Score = 97.4 bits (241), Expect = 3e-21
Identities = 84/304 (27%), Positives = 131/304 (42%), Gaps = 9/304 (2%)
Frame = +2
Query: 33 KELVAGGVAGALSKTAVAPLERVKILWQTRTGGF-----HTLGVCQSLNKLVKHEGFLGL 87
K+L AG V GA P + +K+ Q++ G ++ + + EG GL
Sbjct: 176 KDLAAGTVGGASQLIVGHPFDTIKVKLQSQPTPLPGQPPKFSGAFDAVKQTIAAEGPRGL 355
Query: 88 YKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYP 147
+KG GA + + A+ F T I+ + P + G+ AG P
Sbjct: 356 FKGMGAPLATVAAFNAVLF-TVRGQMETIVRSHPGAPLTVSQQFICGAGAGVAVSFLACP 532
Query: 148 LDLARTKL-AYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTI 206
+L + +L A + G +K P + VL S EGG RGL++G+ PT+
Sbjct: 533 TELIKCRLQAQSALGGSGAAAVAVKYGGPMDV---ARQVLRS---EGGTRGLFKGLVPTM 694
Query: 207 TGILPYAGLKFYTYEKLKMHVPEEHQKSILMR---LSCGALAGLFGQTLTYPLDVVKRQM 263
+P + F YE +K + S L R + G LAG L YP DVVK +
Sbjct: 695 AREIPGNAIMFGVYEAIKQQIAGGTDTSGLGRGSLIVAGGLAGASFWFLVYPTDVVKSVI 874
Query: 264 QVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMK 323
QV +N ++ +LD RKI +G++ L+ G R +P+ A F AY+ +
Sbjct: 875 QVDDYKN-----PKFSGSLDAFRKIQATEGFKGLYKGFGPAMARSIPANAACFLAYEMTR 1039
Query: 324 AWLG 327
+ LG
Sbjct: 1040SALG 1051
>TC7883 homologue to UP|ADT1_GOSHI (O22342) ADP,ATP carrier protein 1,
mitochondrial precursor (ADP/ATP translocase 1) (Adenine
nucleotide translocator 1) (ANT 1), partial (54%)
Length = 856
Score = 96.3 bits (238), Expect = 7e-21
Identities = 67/207 (32%), Positives = 101/207 (48%), Gaps = 9/207 (4%)
Frame = +2
Query: 27 GVPVYVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-------LGVCQSLNKLV 79
G ++ + + GGV+ A+SKTA AP+ERVK+L Q + + G+ + +
Sbjct: 260 GAAGFLADFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKSGRLSEPYKGISDCFGRTM 439
Query: 80 KHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFI--DLLAGSAA 137
K EG + L++GN A+ +R P AL+F + +K G + +L +G AA
Sbjct: 440 KDEGVIALWRGNTANVIRYFPTQALNFAFKDYFKRLFNFKKDKDGYWKWFAGNLASGGAA 619
Query: 138 GGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARG 197
G +S+L Y LD ART+LA D + K G + G+ V + G G
Sbjct: 620 GASSLLFVYSLDYARTRLAN---DAKAAKKGGERQFN------GLIDVYKKTIKSDGIAG 772
Query: 198 LYRGVGPTITGILPYAGLKFYTYEKLK 224
LYRG + GI+ Y GL F Y+ LK
Sbjct: 773 LYRGFNISCVGIIVYRGLYFGMYDSLK 853
>TC9482 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier protein-like
(At5g48970), partial (42%)
Length = 529
Score = 92.0 bits (227), Expect = 1e-19
Identities = 48/149 (32%), Positives = 78/149 (52%), Gaps = 16/149 (10%)
Frame = +3
Query: 194 GARGLYRGVGPTITGILPYAGLKFYTYEKLKM-----------HVPEEHQKSILMRLSCG 242
G +G+Y G+ PT+ I+PYAGL+F TY+ K + E+ S CG
Sbjct: 9 GFQGMYAGLSPTLVEIIPYAGLQFGTYDTFKRWAMAWNHYRYSNTSAENSPSSFQLFLCG 188
Query: 243 ALAGLFGQTLTYPLDVVKRQMQVGSLQN-----AAHEDTRYRNTLDGLRKIVRNQGWRQL 297
AG + + +PLDVVK++ Q+ LQ A E Y + D +++I+R +GW L
Sbjct: 189 LAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYSSMFDAIQRILRLEGWAGL 368
Query: 298 FAGVSINYIRIVPSAAISFTAYDTMKAWL 326
+ G+ + ++ P+ A++F AY+ WL
Sbjct: 369 YKGIIPSTVKAAPAGAVTFVAYELTSDWL 455
Score = 62.0 bits (149), Expect = 1e-10
Identities = 44/147 (29%), Positives = 69/147 (46%), Gaps = 8/147 (5%)
Frame = +3
Query: 83 GFLGLYKGNGASAVRIVPYAALHFMTYERYKSWIL-------NNCPALGSGPFIDL-LAG 134
GF G+Y G + V I+PYA L F TY+ +K W + +N A S L L G
Sbjct: 9 GFQGMYAGLSPTLVEIIPYAGLQFGTYDTFKRWAMAWNHYRYSNTSAENSPSSFQLFLCG 188
Query: 135 SAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGG 194
AAG + L +PLD+ + + +Q+ + + G + R A+ + + + R G
Sbjct: 189 LAAGTCAKLVCHPLDVVKKR--FQIEGLQRHPRYGARVER--RAYSSMFDAIQRILRLEG 356
Query: 195 ARGLYRGVGPTITGILPYAGLKFYTYE 221
GLY+G+ P+ P + F YE
Sbjct: 357 WAGLYKGIIPSTVKAAPAGAVTFVAYE 437
Score = 38.1 bits (87), Expect = 0.002
Identities = 25/92 (27%), Positives = 41/92 (44%), Gaps = 11/92 (11%)
Frame = +3
Query: 36 VAGGVAGALSKTAVAPLERVKILWQTRTGGFHTL-----------GVCQSLNKLVKHEGF 84
+ G AG +K PL+ VK +Q H + ++ ++++ EG+
Sbjct: 180 LCGLAAGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVERRAYSSMFDAIQRILRLEGW 359
Query: 85 LGLYKGNGASAVRIVPYAALHFMTYERYKSWI 116
GLYKG S V+ P A+ F+ YE W+
Sbjct: 360 AGLYKGIIPSTVKAAPAGAVTFVAYELTSDWL 455
Score = 35.8 bits (81), Expect = 0.011
Identities = 13/34 (38%), Positives = 22/34 (64%)
Frame = +3
Query: 292 QGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAW 325
+G++ ++AG+S + I+P A + F YDT K W
Sbjct: 6 RGFQGMYAGLSPTLVEIIPYAGLQFGTYDTFKRW 107
>AV408895
Length = 429
Score = 83.6 bits (205), Expect = 4e-17
Identities = 49/155 (31%), Positives = 77/155 (49%), Gaps = 6/155 (3%)
Frame = +3
Query: 76 NKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSW------ILNNCPALGSGPFI 129
+++V EG +KGN + +PY++++F +YE YK W + N+ + + FI
Sbjct: 9 SRIVNEEGVRAFWKGNLVTIAHRLPYSSVNFYSYEHYKKWLRMVSGVQNHRDNVSADVFI 188
Query: 130 DLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSV 189
+ G AG T+ TYPLDL RT+LA Q T + GI L ++
Sbjct: 189 HFVGGGMAGITAATSTYPLDLVRTRLAAQTNFT---------------YYRGIWHALQTI 323
Query: 190 YREGGARGLYRGVGPTITGILPYAGLKFYTYEKLK 224
+E G GLY+G+G T+ + P + F YE L+
Sbjct: 324 SKEEGILGLYKGLGTTLLTVGPNIAISFSVYESLR 428
Score = 53.5 bits (127), Expect = 5e-08
Identities = 27/75 (36%), Positives = 41/75 (54%)
Frame = +3
Query: 36 VAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASA 95
V GG+AG + T+ PL+ V+ +T + G+ +L + K EG LGLYKG G +
Sbjct: 195 VGGGMAGITAATSTYPLDLVRTRLAAQTNFTYYRGIWHALQTISKEEGILGLYKGLGTTL 374
Query: 96 VRIVPYAALHFMTYE 110
+ + P A+ F YE
Sbjct: 375 LTVGPNIAISFSVYE 419
>TC13252 similar to UP|Q9MA27 (Q9MA27) T5E21.6, partial (17%)
Length = 638
Score = 83.6 bits (205), Expect = 4e-17
Identities = 39/82 (47%), Positives = 58/82 (70%)
Frame = +2
Query: 253 TYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGVSINYIRIVPSA 312
TYPL VV+RQMQV L +A ++ + TL + I GW+QLF+G+SINY+++VPS
Sbjct: 2 TYPLAVVRRQMQVPIL--SASDNVELKGTLRSMVLIAHKHGWKQLFSGLSINYLKVVPSV 175
Query: 313 AISFTAYDTMKAWLGIPPQQKS 334
A+ FT YDTMK +L +P ++++
Sbjct: 176 ALGFTVYDTMKLYLRVPSREEA 241
>TC10512 similar to UP|Q9FI73 (Q9FI73) Mitochondrial carrier protein-like
(At5g48970), partial (43%)
Length = 563
Score = 64.7 bits (156), Expect = 2e-11
Identities = 35/136 (25%), Positives = 68/136 (49%), Gaps = 19/136 (13%)
Frame = +1
Query: 37 AGGVAGALSKTAVAPLERVKILWQTR----------------TGGFHTLGVCQSLNKLVK 80
AG ++G +S+T +PL+ +KI +Q + T G+ Q+ +++
Sbjct: 151 AGAISGGISRTVTSPLDVIKIRFQVQLEPTSSWALLRRNLAATAPSKYTGMFQASKDILR 330
Query: 81 HEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWILNNCPA---LGSGPFIDLLAGSAA 137
EG G ++GN + + ++PY A+ F + K++ + +G P++ ++G+ A
Sbjct: 331 EEGVQGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKTENHIGLSPYLSYVSGALA 510
Query: 138 GGTSVLCTYPLDLART 153
G + L +YP DL RT
Sbjct: 511 GCAATLGSYPFDLLRT 558
Score = 54.3 bits (129), Expect = 3e-08
Identities = 38/140 (27%), Positives = 63/140 (44%), Gaps = 8/140 (5%)
Frame = +1
Query: 129 IDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRG--TIKDGIKGVRPQPAHIGIKSVL 186
ID AG+ +GG S T PLD+ + + Q+ T ++ + P + G+
Sbjct: 139 IDSSAGAISGGISRTVTSPLDVIKIRFQVQLEPTSSWALLRRNLAATAPSK-YTGMFQAS 315
Query: 187 TSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPEEHQKSILMRLS------ 240
+ RE G +G +RG P + ++PY ++F KLK + + LS
Sbjct: 316 KDILREEGVQGFWRGNVPALLMVMPYTAIQFTVLHKLKTFASGSSKTENHIGLSPYLSYV 495
Query: 241 CGALAGLFGQTLTYPLDVVK 260
GALAG +YP D+++
Sbjct: 496 SGALAGCAATLGSYPFDLLR 555
>BP062301
Length = 364
Score = 63.9 bits (154), Expect = 4e-11
Identities = 36/77 (46%), Positives = 49/77 (62%), Gaps = 3/77 (3%)
Frame = +1
Query: 45 SKTAVAPLERVKILWQTRTGGFHTL---GVCQSLNKLVKHEGFLGLYKGNGASAVRIVPY 101
S+TAVAPLER+KIL Q + H + G Q L + + EGF GL+KGNG + RIVP
Sbjct: 106 SRTAVAPLERMKILLQVQNP--HNIKYNGTIQGLKYIWRTEGFRGLFKGNGTNCARIVPN 279
Query: 102 AALHFMTYERYKSWILN 118
+A+ F +YE+ IL+
Sbjct: 280 SAVKFFSYEQASKGILH 330
Score = 34.3 bits (77), Expect = 0.031
Identities = 14/37 (37%), Positives = 24/37 (64%)
Frame = +1
Query: 186 LTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEK 222
L ++R G RGL++G G I+P + +KF++YE+
Sbjct: 199 LKYIWRTEGFRGLFKGNGTNCARIVPNSAVKFFSYEQ 309
>BP039065
Length = 564
Score = 63.9 bits (154), Expect = 4e-11
Identities = 31/84 (36%), Positives = 48/84 (56%), Gaps = 7/84 (8%)
Frame = +3
Query: 37 AGGVAGALSKTAVAPLERVKILWQTR-------TGGFHTLGVCQSLNKLVKHEGFLGLYK 89
AG +AGA +KT APL+R+K+L QT + +G +++ + K EG G +K
Sbjct: 276 AGAIAGAAAKTVTAPLDRIKLLMQTHGVRVGQDSAKKAAIGFVEAITVIGKEEGIRGYWK 455
Query: 90 GNGASAVRIVPYAALHFMTYERYK 113
GN +R++PY+A+ YE YK
Sbjct: 456 GNLPQVIRVIPYSAVQLFAYEIYK 527
Score = 39.3 bits (90), Expect = 0.001
Identities = 25/87 (28%), Positives = 45/87 (50%), Gaps = 3/87 (3%)
Frame = +3
Query: 240 SCGALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTL---DGLRKIVRNQGWRQ 296
S GA+AG +T+T PLD +K MQ ++ +D+ + + + + I + +G R
Sbjct: 273 SAGAIAGAAAKTVTAPLDRIKLLMQTHGVR--VGQDSAKKAAIGFVEAITVIGKEEGIRG 446
Query: 297 LFAGVSINYIRIVPSAAISFTAYDTMK 323
+ G IR++P +A+ AY+ K
Sbjct: 447 YWKGNLPQVIRVIPYSAVQLFAYEIYK 527
>TC9406 similar to UP|Q9M058 (Q9M058) Ca-dependent solute carrier-like
protein, partial (30%)
Length = 676
Score = 63.5 bits (153), Expect = 5e-11
Identities = 39/129 (30%), Positives = 69/129 (53%), Gaps = 5/129 (3%)
Frame = +3
Query: 215 LKFYTYEKLKM----HVPEEHQKSILMRLSCGALAGLFGQTLTYPLDVVKRQMQVGSLQN 270
+ F YE L+ P++ S ++ L+C +L+G+ T+ +PLD+V+R+MQ L+
Sbjct: 12 ISFSVYESLRSLWISRRPDD--SSAVVSLACASLSGVASSTVIFPLDLVRRRMQ---LEG 176
Query: 271 AAHEDTRYRNTLDG-LRKIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAWLGIP 329
Y + G + I++N+G R L+ G+ Y ++VP I+F Y+T+K L
Sbjct: 177 VGGRACVYNTGMCGAFQCIIKNEGVRGLYRGILPEYYKVVPGVGIAFMTYETLKRLLSST 356
Query: 330 PQQKSRSVS 338
P + SV+
Sbjct: 357 PS*LAYSVA 383
Score = 53.5 bits (127), Expect = 5e-08
Identities = 34/122 (27%), Positives = 54/122 (43%)
Frame = +3
Query: 103 ALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDT 162
A+ F YE +S ++ P S + L S +G S +PLDL R ++ + +
Sbjct: 9 AISFSVYESLRSLWISRRPD-DSSAVVSLACASLSGVASSTVIFPLDLVRRRMQLEGVGG 185
Query: 163 RGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEK 222
R + + G+ + + G RGLYRG+ P ++P G+ F TYE
Sbjct: 186 RACVYN-----------TGMCGAFQCIIKNEGVRGLYRGILPEYYKVVPGVGIAFMTYET 332
Query: 223 LK 224
LK
Sbjct: 333 LK 338
Score = 48.9 bits (115), Expect = 1e-06
Identities = 29/95 (30%), Positives = 48/95 (50%), Gaps = 4/95 (4%)
Frame = +3
Query: 32 VKELVAGGVAGALSKTAVAPLERVKILWQTRTGG----FHTLGVCQSLNKLVKHEGFLGL 87
V L ++G S T + PL+ V+ Q G + G+C + ++K+EG GL
Sbjct: 81 VVSLACASLSGVASSTVIFPLDLVRRRMQLEGVGGRACVYNTGMCGAFQCIIKNEGVRGL 260
Query: 88 YKGNGASAVRIVPYAALHFMTYERYKSWILNNCPA 122
Y+G ++VP + FMTYE K +L++ P+
Sbjct: 261 YRGILPEYYKVVPGVGIAFMTYETLKR-LLSSTPS 362
>BP071178
Length = 408
Score = 63.5 bits (153), Expect = 5e-11
Identities = 40/128 (31%), Positives = 69/128 (53%), Gaps = 2/128 (1%)
Frame = +2
Query: 112 YKSWILNNC--PALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDG 169
YKS + N L SG + +AG+AAG TS++ YPLD+A T+LA + T
Sbjct: 53 YKSMLRGNSYDDNLFSGASANFVAGAAAGCTSLVLVYPLDIAHTRLAADIGRTE------ 214
Query: 170 IKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKMHVPE 229
++ R GI L +++ + G RG+YRG+ ++ G++ + GL F ++ +K + E
Sbjct: 215 VRQFR------GIHHFLVTIFHKDGIRGIYRGLPASLHGMVVHRGLYFGGFDTIKEMLTE 376
Query: 230 EHQKSILM 237
E + + +
Sbjct: 377 ESEPEVAL 400
Score = 30.8 bits (68), Expect = 0.34
Identities = 22/85 (25%), Positives = 36/85 (41%)
Frame = +2
Query: 242 GALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRNQGWRQLFAGV 301
GA AG L YPLD+ ++ + E ++R L I G R ++ G+
Sbjct: 125 GAAAGCTSLVLVYPLDIAHTRL---AADIGRTEVRQFRGIHHFLVTIFHKDGIRGIYRGL 295
Query: 302 SINYIRIVPSAAISFTAYDTMKAWL 326
+ +V + F +DT+K L
Sbjct: 296 PASLHGMVVHRGLYFGGFDTIKEML 370
>CB827990
Length = 547
Score = 62.8 bits (151), Expect = 8e-11
Identities = 51/157 (32%), Positives = 77/157 (48%), Gaps = 22/157 (14%)
Frame = +3
Query: 194 GARGLYRGVGPTITGILPYAGLKFYTYEKLK---MHVPE-----EHQKSILMRLSC---- 241
G LY+G+ P+I + P + + Y+ LK +H PE +H K L+
Sbjct: 18 GFFSLYKGLVPSIVSMAPSGAVFYGVYDILKSAYLHSPEGMKRIQHMKEETEELNAFDQL 197
Query: 242 ----------GALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLRKIVRN 291
GA+AG + TYP +VV+RQ+Q +Q A TR NTL KIV
Sbjct: 198 ELGPVRTLLYGAIAGCCAEAATYPFEVVRRQLQ---MQVRA---TRL-NTLATCVKIVEQ 356
Query: 292 QGWRQLFAGVSINYIRIVPSAAISFTAYDTMKAWLGI 328
G +AG+ + ++++PSAAIS+ Y+ MK L +
Sbjct: 357 GGVPAFYAGLIPSLLQVLPSAAISYFVYEFMKIVLKV 467
Score = 62.4 bits (150), Expect = 1e-10
Identities = 45/167 (26%), Positives = 69/167 (40%), Gaps = 19/167 (11%)
Frame = +3
Query: 78 LVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWI-------------------LN 118
++K EGF LYKG S V + P A+ + Y+ KS LN
Sbjct: 3 MIKTEGFFSLYKGLVPSIVSMAPSGAVFYGVYDILKSAYLHSPEGMKRIQHMKEETEELN 182
Query: 119 NCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIKDGIKGVRPQPA 178
L GP LL G+ AG + TYP ++ R +L QV TR
Sbjct: 183 AFDQLELGPVRTLLYGAIAGCCAEAATYPFEVVRRQLQMQVRATR--------------- 317
Query: 179 HIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLKM 225
+ + + +GG Y G+ P++ +LP A + ++ YE +K+
Sbjct: 318 -LNTLATCVKIVEQGGVPAFYAGLIPSLLQVLPSAAISYFVYEFMKI 455
>CB828090
Length = 427
Score = 61.6 bits (148), Expect = 2e-10
Identities = 28/87 (32%), Positives = 50/87 (57%), Gaps = 5/87 (5%)
Frame = +2
Query: 245 AGLFGQTLTYPLDVVKRQMQVGSLQN-----AAHEDTRYRNTLDGLRKIVRNQGWRQLFA 299
AG + + +PLDVVK++ Q+ LQ A E Y+N D +++I++ +GW L+
Sbjct: 5 AGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAYKNMFDAMKRIIQMEGWAGLYK 184
Query: 300 GVSINYIRIVPSAAISFTAYDTMKAWL 326
G+ + ++ P+ A++F AY+ WL
Sbjct: 185 GLFPSTVKAAPAGAVTFVAYELTSDWL 265
Score = 38.1 bits (87), Expect = 0.002
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 11/87 (12%)
Frame = +2
Query: 41 AGALSKTAVAPLERVKILWQTRTGGFHTL-----------GVCQSLNKLVKHEGFLGLYK 89
AG +K PL+ VK +Q H + ++ ++++ EG+ GLYK
Sbjct: 5 AGTCAKLVCHPLDVVKKRFQIEGLQRHPRYGARVEHRAYKNMFDAMKRIIQMEGWAGLYK 184
Query: 90 GNGASAVRIVPYAALHFMTYERYKSWI 116
G S V+ P A+ F+ YE W+
Sbjct: 185 GLFPSTVKAAPAGAVTFVAYELTSDWL 265
>TC19374 similar to UP|Q8LNZ1 (Q8LNZ1) Mitochondrial uncoupling protein,
partial (49%)
Length = 556
Score = 55.8 bits (133), Expect = 1e-08
Identities = 42/130 (32%), Positives = 65/130 (49%), Gaps = 8/130 (6%)
Frame = +3
Query: 143 LCTYPLDLARTKLAYQVIDTRGTIKDGIKG-VRPQPAHIGIKSVLTSVYREGGARGLYRG 201
+CT PLD A+ +L Q K GI G V P + G+ + ++ RE GA L++G
Sbjct: 165 VCTIPLDTAKVRLQLQ--------KQGIAGDVASLPKYKGMLGTIATIAREEGASALWKG 320
Query: 202 VGPTITGILPYAGLKFYTYEKLK-MHVPEEH------QKSILMRLSCGALAGLFGQTLTY 254
+ P + Y GL+ YE +K ++V +H K IL + GA+A T+
Sbjct: 321 IVPGLHRQCLYGGLRIGLYEPVKALYVGSDHVGDVPLSKKILAAFTTGAVA----ITVAN 488
Query: 255 PLDVVKRQMQ 264
P D+VK ++Q
Sbjct: 489 PTDLVKVRLQ 518
>TC7843 weakly similar to GB|AAF03412.1|6090963|AF188712 mitochondrial
dicarboxylate carrier {Mus musculus;} , partial (22%)
Length = 1594
Score = 54.7 bits (130), Expect = 2e-08
Identities = 41/191 (21%), Positives = 82/191 (42%), Gaps = 1/191 (0%)
Frame = +3
Query: 35 LVAGGVAGALSKTAVAPLERVKILWQTRTGGFHTLG-VCQSLNKLVKHEGFLGLYKGNGA 93
L+AGG+ A+ A + R++ + V ++ ++ K EG L++G+
Sbjct: 663 LIAGGIGAAVGNPADVAMVRMQADGRLPAAQRRNYSSVVDAITRMAKQEGVTSLWRGSSL 842
Query: 94 SAVRIVPYAALHFMTYERYKSWILNNCPALGSGPFIDLLAGSAAGGTSVLCTYPLDLART 153
+ R + A +Y+++K IL + G + A AAG + + + P+D+ +T
Sbjct: 843 TVNRAMLVTASQLASYDQFKEMILEK-GLMRDGLGTHVTASFAAGFVAAVASNPVDVIKT 1019
Query: 154 KLAYQVIDTRGTIKDGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYA 213
++ ++ +P + G R G LY+G PTI+ P+
Sbjct: 1020RVMNMKVEPGA-----------EPPYAGALDCALKTVRAEGPMALYKGFIPTISRQGPFT 1166
Query: 214 GLKFYTYEKLK 224
+ F T E+++
Sbjct: 1167VVLFVTLEQVR 1199
Score = 30.0 bits (66), Expect = 0.58
Identities = 28/130 (21%), Positives = 48/130 (36%), Gaps = 32/130 (24%)
Frame = +3
Query: 242 GALAGLFGQTLTYPLDVVKRQMQVGSLQNAAHEDTRYRNTLDGLR--------------- 286
G +A + T+PLD++K +MQ+ NAA + + +
Sbjct: 270 GGIASIIAGCSTHPLDLIKVRMQLQGEANAAPKPVQQLRPALAFQNGSTIHVAAHPHSPP 449
Query: 287 ----------------KIVRNQGWRQLFAGVSINYIRIVPSAAISFTAYDTMK-AWLGIP 329
++V+ +G R LF+GVS +R + + YD +K W
Sbjct: 450 PPPMPQPRVGPISVGVRLVQQEGLRALFSGVSATVLRQMLYSTTRMGLYDILKQKWSNGG 629
Query: 330 PQQKSRSVSA 339
SR + A
Sbjct: 630 NMPLSRKIEA 659
>TC11705 homologue to UP|O49875 (O49875) Adenine nucleotide translocator,
partial (38%)
Length = 580
Score = 52.4 bits (124), Expect = 1e-07
Identities = 30/90 (33%), Positives = 49/90 (54%), Gaps = 7/90 (7%)
Frame = +3
Query: 31 YVKELVAGGVAGALSKTAVAPLERVKILWQTRTGGFHT-------LGVCQSLNKLVKHEG 83
++ + + GGV+ A+SKTA AP+ERVK+L Q + G+ + +K EG
Sbjct: 270 FLIDFLMGGVSAAVSKTAAAPIERVKLLIQNQDEMIKAGRLSEPYKGIGDCFTRTMKDEG 449
Query: 84 FLGLYKGNGASAVRIVPYAALHFMTYERYK 113
+ L +GN A+ +R P AL+F + +K
Sbjct: 450 VVAL*RGNTANVIRYFPTQALNFAFKDYFK 539
>TC17966 similar to UP|Q8L9F8 (Q8L9F8) Mitochondrial carrier-like protein,
partial (53%)
Length = 917
Score = 50.1 bits (118), Expect = 5e-07
Identities = 60/237 (25%), Positives = 96/237 (40%), Gaps = 14/237 (5%)
Frame = +2
Query: 2 DSSQGSTLAGLVDNASIQKDQSCFDGVPVYVKELVAGGVAGALSKTAVAPLERVKILWQT 61
D++ + + AS Q + + V + L+ G G SKT +A L ++
Sbjct: 47 DTTSTAVASAAAGVASAMAAQLVWTPIDVVSQRLMVQGTGG--SKTMLANLNS-----ES 205
Query: 62 RTGGFHTLGVCQSLNKLVKHEGFLGLYKGNGASAVRIVPYAALHFMTYERYKSWIL---- 117
GF + K++ +G G Y+G G S + P A+ + +Y +I
Sbjct: 206 YRNGF------DAFRKILCADGPKGFYRGFGISLLTYAPSNAVWWTSYSMVHRFIWGSFG 367
Query: 118 -------NNC---PALGSGPFIDLLAGSAAGGTSVLCTYPLDLARTKLAYQVIDTRGTIK 167
N C P S + L+ A G S + T PLD +T+L QV+DT
Sbjct: 368 PYMGQRDNVCGFRPDSKSIVAVQGLSAVVASGVSAIVTMPLDTIQTRL--QVLDTE---- 529
Query: 168 DGIKGVRPQPAHIGIKSVLTSVYREGGARGLYRGVGPTITGILPYAGLKFYTYEKLK 224
+ R +P + L ++ +EGG YRG+GP + A TYE +K
Sbjct: 530 ---ENGRRRP--LTSVQTLRNLVKEGGLLACYRGLGPRWASMSMSATTMITTYEFVK 685
Score = 40.4 bits (93), Expect = 4e-04
Identities = 25/92 (27%), Positives = 47/92 (50%), Gaps = 5/92 (5%)
Frame = +2
Query: 240 SCGALAGLFGQTLTYPLDVVKRQMQV----GSLQNAAHEDTR-YRNTLDGLRKIVRNQGW 294
+ G + + Q + P+DVV +++ V GS A+ ++ YRN D RKI+ G
Sbjct: 77 AAGVASAMAAQLVWTPIDVVSQRLMVQGTGGSKTMLANLNSESYRNGFDAFRKILCADGP 256
Query: 295 RQLFAGVSINYIRIVPSAAISFTAYDTMKAWL 326
+ + G I+ + PS A+ +T+Y + ++
Sbjct: 257 KGFYRGFGISLLTYAPSNAVWWTSYSMVHRFI 352
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,515,784
Number of Sequences: 28460
Number of extensions: 69106
Number of successful extensions: 402
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 319
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 365
length of query: 340
length of database: 4,897,600
effective HSP length: 91
effective length of query: 249
effective length of database: 2,307,740
effective search space: 574627260
effective search space used: 574627260
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0349b.8


