

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
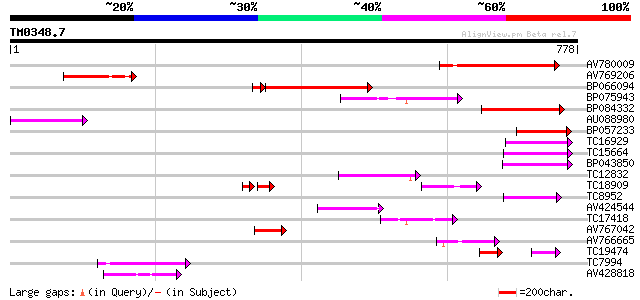
Query= TM0348.7
(778 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV780009 151 3e-37
AV769206 142 3e-34
BP066094 114 5e-28
BP075943 102 3e-22
BP084332 89 3e-18
AU088980 87 1e-17
BP057233 79 2e-15
TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%) 73 2e-13
TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, p... 67 8e-12
BP043850 66 2e-11
TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 63 2e-10
TC18909 61 8e-10
TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%) 60 1e-09
AV424544 58 5e-09
TC17418 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partia... 53 2e-07
AV767042 52 5e-07
AV766665 51 6e-07
TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retro... 37 2e-05
TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomeras... 43 2e-04
AV428818 42 4e-04
>AV780009
Length = 529
Score = 151 bits (382), Expect = 3e-37
Identities = 79/166 (47%), Positives = 104/166 (62%), Gaps = 1/166 (0%)
Frame = -1
Query: 590 KAVKRILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGSCVFLGPNL 649
+A R+LRY+KG G+ S PL L A+ D+DW PD RRS +G +FLG +L
Sbjct: 529 QAATRVLRYVKGAPAQGLFF---SADSPLKLQAYSDSDWAGCPDTRRSVTGYSIFLGTSL 359
Query: 650 ISWTAKKQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVL-CDNMSTVLL 708
ISW KKQT V+RSS+EAEYR+LA T E+ W+ L LK+ +P L CDN S + +
Sbjct: 358 ISWRTKKQTTVSRSSSEAEYRALAATVCEVQWLSYLFQFLKLNVPLPVPLFCDNQSALHI 179
Query: 709 THNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTK 754
HNP H RTKH+E+D VR K+QA + + + + HQ ADIFTK
Sbjct: 178 AHNPTFHERTKHIELDCHVVRAKLQAGLIHLLPISTHHQLADIFTK 41
>AV769206
Length = 536
Score = 142 bits (357), Expect = 3e-34
Identities = 69/100 (69%), Positives = 82/100 (82%)
Frame = -3
Query: 75 FLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLDQSGRIYVSKDVLFHEHRFPYTTLFPSE 134
FLRPYNS KLSLHSKECVF+GYS +HKGYKCLDQSGRIY+SKDVLFHEHRFPY +LFP E
Sbjct: 534 FLRPYNSTKLSLHSKECVFIGYSINHKGYKCLDQSGRIYISKDVLFHEHRFPYPSLFPDE 355
Query: 135 PFSPPTSSAEYFPLSTVPIISRSMPQPSPAPISTELANPG 174
S ++SAE+FPLST+PI+S + P PA S+ ++ G
Sbjct: 354 TTS--STSAEFFPLSTIPIVSHTTP---PALASSGFSSLG 250
>BP066094
Length = 532
Score = 114 bits (285), Expect(2) = 5e-28
Identities = 60/148 (40%), Positives = 90/148 (60%), Gaps = 1/148 (0%)
Frame = +2
Query: 351 KPITVRLILSLAISRGWPLQQIDVNNAFLNGVLEEEVYMTQPPGFE-HKDKTLVCKLHKA 409
K +RL++S +++ L Q++V +AFLNG + EEVY+ QPPG E K+ + KL K+
Sbjct: 89 KTEAIRLLISFSVNHNIILHQMNVKSAFLNGYISEEVYVHQPPGXEDEKNSDHIFKLKKS 268
Query: 410 LYGLKQAPRAWFHRLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMV 469
LYGLKQAPRAW+ RL LL+ K D +LF + + IYVDDII +
Sbjct: 269 LYGLKQAPRAWYERLSSFLLENEXVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANPS 448
Query: 470 LIQQLTAKLHSIFALKQLGQLDYFLGVQ 497
L ++ + + + F ++ +G+L YFLG+Q
Sbjct: 449 LCKEFSEMMQAEFEMRMMGELKYFLGIQ 532
Score = 27.7 bits (60), Expect(2) = 5e-28
Identities = 11/18 (61%), Positives = 14/18 (77%)
Frame = +3
Query: 334 YSQVQGFDYSETFSPVVK 351
YSQ G DY+ETF+PV +
Sbjct: 39 YSQQ*GIDYTETFAPVAR 92
>BP075943
Length = 547
Score = 102 bits (253), Expect = 3e-22
Identities = 64/178 (35%), Positives = 94/178 (51%), Gaps = 10/178 (5%)
Frame = -3
Query: 454 MLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQVTHLANGSLLLNQTKY 513
+L+YVDD++L G+ M IQ + + LH F +K LG+ YFLG+++ +G ++LNQ KY
Sbjct: 521 LLLYVDDVLLAGNDMHEIQLVKSSLHDQFRIKDLGEAKYFLGLEIARSTSG-IVLNQRKY 345
Query: 514 INDLLTKVNMADAAPISTPMQFGAKLTKHG----------GTSLHDPTEYRSVVGALQYA 563
L++ S F + HG GT L D YR +VG L Y
Sbjct: 344 ALQLISD---------SGHFGFQPRFYSHGTNSQTLGTNTGTPLTDIGSYRRIVGRLLYL 192
Query: 564 TITRPEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSMTQPLPLL 621
TRP+I++AVN++ QFLS P + H + + L+YL G+ G+ S T LP L
Sbjct: 191 NTTRPDITFAVNQLSQFLSAPTDIHEQQLTGFLKYL*GSPGSGLFYPASSSTL*LPSL 18
>BP084332
Length = 368
Score = 88.6 bits (218), Expect = 3e-18
Identities = 46/115 (40%), Positives = 73/115 (63%), Gaps = 1/115 (0%)
Frame = +2
Query: 648 NLISWTAK-KQTLVARSSTEAEYRSLANTTAELLWVESLLTELKIAFTVPTVLCDNMSTV 706
N+ W A Q+ +A S+ EAEY S A + ++LW++ L + +I + + CDN + +
Sbjct: 2 NVNFWEAT*GQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAI 181
Query: 707 LLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKALSPTRF 761
L+ NPILH+R KH+E+ F+R+ VQ L+++ V ++HQ ADIFTK L+ RF
Sbjct: 182 SLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRF 346
>AU088980
Length = 360
Score = 86.7 bits (213), Expect = 1e-17
Identities = 41/106 (38%), Positives = 60/106 (55%)
Frame = +2
Query: 2 VERKHRHIVETGLALLSHASMPLHFWDHAFLTATYLINRMSTTTLQGASPYFKLYGNHPD 61
VERKHRHI+ AL+ H+ +P + W A A YLIN + + L+G PY L + P
Sbjct: 11 VERKHRHILNVTRALMFHSYLPKNLWTFAVKHAAYLINXLPSPLLKGQCPYQFLNNDSPT 190
Query: 62 FKSLKIFGSACFPFLRPYNSNKLSLHSKECVFLGYSSSHKGYKCLD 107
LK+FG+ C+ +N KL +++CVFLG+ + GY +D
Sbjct: 191 LLDLKVFGTLCYSSTLTHNXQKLDPRARKCVFLGFKTGTXGYIVMD 328
>BP057233
Length = 473
Score = 79.3 bits (194), Expect = 2e-15
Identities = 36/76 (47%), Positives = 52/76 (68%)
Frame = -2
Query: 696 PTVLCDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKA 755
P + CDN+S L NP+LH R+KH+E+D+ ++R++V +VV +VP+ Q AD TK
Sbjct: 451 PILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNEVVVAYVPTTDQIADCLTKP 272
Query: 756 LSPTRFLLMRDKLNVV 771
LS TRF +RDKL V+
Sbjct: 271 LSHTRFSQLRDKLGVI 224
>TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%)
Length = 553
Score = 73.2 bits (178), Expect = 2e-13
Identities = 38/93 (40%), Positives = 55/93 (58%), Gaps = 1/93 (1%)
Frame = +1
Query: 681 WVESLLTELKIAFTVPT-VLCDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKSLVV 739
W+ LL +LK+ F P V CDN S + NP+ H RTKH+E+D VRE++Q + +
Sbjct: 4 WLTYLLQDLKVPFEQPALVYCDNNSARHIAANPVFHERTKHIEIDCHIVRERIQKGLIHL 183
Query: 740 QHVPSEHQRADIFTKALSPTRFLLMRDKLNVVD 772
+ S ADI+TKALSP F + KL +++
Sbjct: 184 LPISSSEPLADIYTKALSPQNFHQICAKLGLIN 282
>TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, partial (4%)
Length = 670
Score = 67.4 bits (163), Expect = 8e-12
Identities = 37/96 (38%), Positives = 56/96 (57%), Gaps = 1/96 (1%)
Frame = +1
Query: 678 ELLWVESLLTELKIAFTVPTVL-CDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKS 736
EL W+ +L +L++ F P++L CD+ S + N + H RTKH+++D VREK+QAK
Sbjct: 4 EL*WLTYILQDLRVPFISPSLLYCDSQSARHIATNAVFHERTKHLDIDCHVVREKLQAKL 183
Query: 737 LVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNVVD 772
+ + S Q ADI TK L F + KL V++
Sbjct: 184 FHLLPISSVDQTADILTKPLESGPFSHLVSKLGVLN 291
>BP043850
Length = 515
Score = 65.9 bits (159), Expect = 2e-11
Identities = 35/97 (36%), Positives = 56/97 (57%), Gaps = 1/97 (1%)
Frame = -1
Query: 677 AELLWVESLLTELKIAFTVPTVL-CDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAK 735
+E++W+ LL+EL + PT L DN S + + NP+ H T+H+E+D VRE +
Sbjct: 515 SEIIWLRGLLSELGFLQSQPTPLHADNTSAIQIAANPVYHEWTRHIEVDCHSVREAYDRR 336
Query: 736 SLVVQHVPSEHQRADIFTKALSPTRFLLMRDKLNVVD 772
+ + HV + Q ADI TK+L+ R + KL +V+
Sbjct: 335 VITLPHVSTSVQIADILTKSLTRQRHNFLVSKLMLVE 225
>TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (9%)
Length = 747
Score = 63.2 bits (152), Expect = 2e-10
Identities = 40/119 (33%), Positives = 62/119 (51%), Gaps = 7/119 (5%)
Frame = +2
Query: 452 IYMLIYVDDIILTGDSMVLIQQLTAKLHSIFALKQLGQLDYFLGVQV-THLANGSLLLNQ 510
I +L+YVDD+++ G + +Q+L A+L F +K LG + LG+Q+ + + L+Q
Sbjct: 89 IILLLYVDDMLVVGPNKDRVQELKAQLAREFDMKDLGPANKILGMQIHRDRKDRRIWLSQ 268
Query: 511 TKYINDLLTKVNMADAAPISTPMQFGAKLTKHGGTSLH------DPTEYRSVVGALQYA 563
Y+ +L + NM D PISTP+ KL+ S Y S VG+L YA
Sbjct: 269 KNYLQKVLRRFNMQDCNPISTPLPVNYKLSSSMIPSSEAERMEMSRVPYASAVGSLMYA 445
>TC18909
Length = 621
Score = 60.8 bits (146), Expect = 8e-10
Identities = 41/85 (48%), Positives = 48/85 (56%), Gaps = 2/85 (2%)
Frame = +2
Query: 565 ITRPEISYAVNKVCQFLSDPHEEHWKAVKRILRYL-KGTITHGVLLQPCSMTQPLPLLAF 623
+T P+IS++VNKVC FLS+ EEH A K IL K T G +Q AF
Sbjct: 323 LTSPQISFSVNKVC*FLSESLEEHGTAGKCILNVT*KAL*TMGFFIQ-------TNFPAF 481
Query: 624 CDADWGSDPDDRRSTSGSCVF-LGP 647
DADW S+ DDRRST VF LGP
Sbjct: 482 SDADWASNVDDRRSTFWEIVFTLGP 556
Score = 40.8 bits (94), Expect(2) = 4e-08
Identities = 17/23 (73%), Positives = 22/23 (94%)
Frame = +2
Query: 341 DYSETFSPVVKPITVRLILSLAI 363
DY+ET SPVVKP+TVR++LSLA+
Sbjct: 62 DYTETVSPVVKPVTVRILLSLAV 130
Score = 33.9 bits (76), Expect(2) = 4e-08
Identities = 15/17 (88%), Positives = 16/17 (93%)
Frame = +1
Query: 320 GSINKYKARLVAKGYSQ 336
GSINKYKARLVAKG+ Q
Sbjct: 1 GSINKYKARLVAKGFHQ 51
>TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%)
Length = 550
Score = 60.5 bits (145), Expect = 1e-09
Identities = 34/81 (41%), Positives = 48/81 (58%), Gaps = 1/81 (1%)
Frame = +3
Query: 678 ELLWVESLLTELKIAFTVPTVL-CDNMSTVLLTHNPILHTRTKHMEMDLFFVREKVQAKS 736
E LW+ L +L+IA + V+ CDN S + L N + H RT+++E+D V KV
Sbjct: 21 EALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFGI 200
Query: 737 LVVQHVPSEHQRADIFTKALS 757
L + HVPS Q AD+FTK +S
Sbjct: 201 LHLLHVPSSDQVADVFTKTIS 263
>AV424544
Length = 276
Score = 58.2 bits (139), Expect = 5e-09
Identities = 37/91 (40%), Positives = 50/91 (54%)
Frame = +3
Query: 423 RLKEVLLQFGFKASKCDPSLFTYNSPLGCIYMLIYVDDIILTGDSMVLIQQLTAKLHSIF 482
+L L G+ S D SLFT +L+YVDD+IL G+ + IQ + KL F
Sbjct: 6 KLSSYLHILGYIQSAHDHSLFTKFRDASFTVILVYVDDLILAGNDLNEIQCVKNKLDIQF 185
Query: 483 ALKQLGQLDYFLGVQVTHLANGSLLLNQTKY 513
+K LG L YFLG++V + G L L+Q KY
Sbjct: 186 RIKDLGTLKYFLGLEVARSSCG-LFLSQRKY 275
>TC17418 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partial (20%)
Length = 739
Score = 52.8 bits (125), Expect = 2e-07
Identities = 34/108 (31%), Positives = 59/108 (54%), Gaps = 3/108 (2%)
Frame = -1
Query: 510 QTKYINDLLTKVNMADAAPISTPMQFGAKLTKHG---GTSLHDPTEYRSVVGALQYATIT 566
Q Y++D+L K M ++ IST + G K + G G D T Y+S++G+++Y
Sbjct: 739 QKIYVDDILKKFKMTNSKYISTTI--GGKEIEAGRRNGGKRVDSTYYKSLIGSVRYLNTV 566
Query: 567 RPEISYAVNKVCQFLSDPHEEHWKAVKRILRYLKGTITHGVLLQPCSM 614
R +I V +F+ +P + H + +R LRY+KGT+ G+ + +M
Sbjct: 565 RSDIVCGVGLRSRFM-EP*DCH*QGAQRSLRYIKGTLKDGIFMLVITM 425
>AV767042
Length = 444
Score = 51.6 bits (122), Expect = 5e-07
Identities = 20/44 (45%), Positives = 32/44 (72%)
Frame = -3
Query: 336 QVQGFDYSETFSPVVKPITVRLILSLAISRGWPLQQIDVNNAFL 379
Q G D ++TFSPVVKP +R + ++A+SR WP+ Q++V N ++
Sbjct: 349 QTAGVDCNKTFSPVVKPAPIRTVFTIALSRSWPIPQLNVQNLWI 218
>AV766665
Length = 601
Score = 51.2 bits (121), Expect = 6e-07
Identities = 33/90 (36%), Positives = 47/90 (51%), Gaps = 4/90 (4%)
Frame = +3
Query: 586 EEHWKAVK----RILRYLKGTITHGVLLQPCSMTQPLPLLAFCDADWGSDPDDRRSTSGS 641
+ W ++K RI YLK G L Q + + F AD+ DR ST G
Sbjct: 336 KNRWSSLKTFTTRIF*YLKANSRRGPLFQKEGKSS---MDGFTYADYLGSIVDRLSTMGY 506
Query: 642 CVFLGPNLISWTAKKQTLVARSSTEAEYRS 671
+FL NL++W +K+Q ++ARSS EAE R+
Sbjct: 507 YMFLSGNLVTWRSKQQNIIARSSGEAELRA 596
>TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retroelement pol
polyprotein, partial (3%)
Length = 517
Score = 37.4 bits (85), Expect(2) = 2e-05
Identities = 17/32 (53%), Positives = 22/32 (68%)
Frame = -1
Query: 645 LGPNLISWTAKKQTLVARSSTEAEYRSLANTT 676
L NLISW +K+ +VARS EAE+R +A T
Sbjct: 517 LSDNLISWKSKETIIVARSRAEAEFRVMATAT 422
Score = 28.5 bits (62), Expect(2) = 2e-05
Identities = 13/39 (33%), Positives = 22/39 (56%)
Frame = -2
Query: 717 RTKHMEMDLFFVREKVQAKSLVVQHVPSEHQRADIFTKA 755
R KH+E+D F+ ++ + + V S Q D+FTK+
Sbjct: 309 RAKHIEIDFHFL*DRRLYLDISTRFVNSNDQLTDVFTKS 193
>TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomerase
precursor , partial (81%)
Length = 1355
Score = 43.1 bits (100), Expect = 2e-04
Identities = 41/134 (30%), Positives = 57/134 (41%), Gaps = 7/134 (5%)
Frame = +2
Query: 121 HEHRFPYTTLFPSEPFSPPTSS-AEYFPLSTVP--IISRSMPQPSPAPISTELANPGPLS 177
H H P T P PPT S A PL+ P + + S+P PSP S + P +
Sbjct: 278 HHHLSPPHTTMP-----PPTLSYAPPLPLNYAPPPLPNPSVPSPSPKTTSRDSPPTRPST 442
Query: 178 PQSEASDLQSQPSPIPTGS--GLASTSQPAEHASSESAHQEMATSS--GVHAASSASTVA 233
S A S P+P P S AS S PA +S ++ A S A+ S S+
Sbjct: 443 TSSPAWSSASAPAPPPPSSSPSSASFSPPANSPTSSASPPPSARRSRRDPSASRSPSSTT 622
Query: 234 VPVNAHPMQTRSKS 247
+P + P ++S
Sbjct: 623 IPASISPSTAPTRS 664
Score = 31.2 bits (69), Expect = 0.66
Identities = 29/95 (30%), Positives = 36/95 (37%), Gaps = 18/95 (18%)
Frame = +2
Query: 126 PYTTLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPSPAPIS------TELANPGPL--- 176
P TT S P P T+S+ + ++ P PS A S T A+P P
Sbjct: 398 PKTTSRDSPPTRPSTTSSPAWSSASAPAPPPPSSSPSSASFSPPANSPTSSASPPPSARR 577
Query: 177 ---------SPQSEASDLQSQPSPIPTGSGLASTS 202
SP S PS PT S L+STS
Sbjct: 578 SRRDPSASRSPSSTTIPASISPSTAPTRSTLSSTS 682
>AV428818
Length = 417
Score = 42.0 bits (97), Expect = 4e-04
Identities = 33/109 (30%), Positives = 53/109 (48%), Gaps = 2/109 (1%)
Frame = +3
Query: 129 TLFPSEPFSPPTSSAEYFPL--STVPIISRSMPQPSPAPISTELANPGPLSPQSEASDLQ 186
TL + P S P + + P S P + S P+P+ ST A P LSP++ S
Sbjct: 69 TLSAATPPSSPQTHQRH*PQRRSQEPPSTSSNPKPTRNASSTSAAPP--LSPRTSTSTAS 242
Query: 187 SQPSPIPTGSGLASTSQPAEHASSESAHQEMATSSGVHAASSASTVAVP 235
PSP P S A+TS P+ ++S S + +++ V + + + + A P
Sbjct: 243 PSPSPSP-NSKTATTSPPSTNSSGTSPPSALTSATSVSSRNPSCSTARP 386
Score = 30.0 bits (66), Expect = 1.5
Identities = 24/86 (27%), Positives = 37/86 (42%)
Frame = +3
Query: 126 PYTTLFPSEPFSPPTSSAEYFPLSTVPIISRSMPQPSPAPISTELANPGPLSPQSEASDL 185
P +T +P +S++ PLS S + P PSP+P S SP S S
Sbjct: 144 PPSTSSNPKPTRNASSTSAAPPLSPRTSTSTASPSPSPSPNS----KTATTSPPSTNSSG 311
Query: 186 QSQPSPIPTGSGLASTSQPAEHASSE 211
S PS + + + ++S + A E
Sbjct: 312 TSPPSALTSATSVSSRNPSCSTARPE 389
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,473,256
Number of Sequences: 28460
Number of extensions: 236953
Number of successful extensions: 2413
Number of sequences better than 10.0: 315
Number of HSP's better than 10.0 without gapping: 2006
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2231
length of query: 778
length of database: 4,897,600
effective HSP length: 97
effective length of query: 681
effective length of database: 2,136,980
effective search space: 1455283380
effective search space used: 1455283380
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0348.7


