

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0347b.6
(1014 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
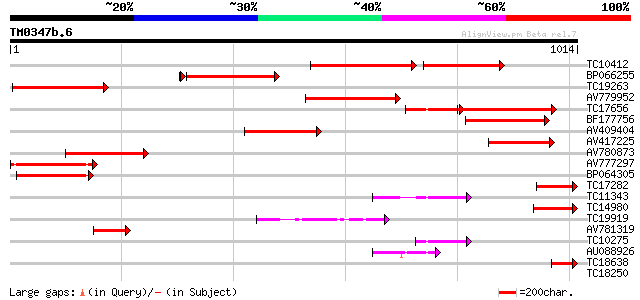
Score E
Sequences producing significant alignments: (bits) Value
TC10412 homologue to UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATP... 313 e-159
BP066255 297 3e-81
TC19263 similar to UP|Q8L8A0 (Q8L8A0) Type IIB calcium ATPase MC... 275 3e-74
AV779952 250 9e-67
TC17656 similar to GB|BAA97361.1|8843813|AB023042 Ca2+-transport... 213 1e-55
BF177756 204 7e-53
AV409404 166 2e-41
AV417225 144 9e-35
AV780873 127 1e-29
AV777297 113 1e-25
BP064305 112 3e-25
TC17282 similar to UP|Q8L8A0 (Q8L8A0) Type IIB calcium ATPase MC... 108 3e-24
TC11343 similar to PIR|B96660|B96660 protein F2K11.18 [imported]... 100 1e-21
TC14980 similar to UP|ACA9_ARATH (Q9LU41) Potential calcium-tran... 69 3e-12
TC19919 homologue to UP|Q7Y068 (Q7Y068) Plasma membrane H+-ATPas... 64 1e-10
AV781319 59 3e-09
TC10275 similar to UP|Q7Y051 (Q7Y051) Paa2 P-type ATPase, partia... 55 6e-08
AU088926 44 1e-04
TC18638 similar to UP|Q8W0V0 (Q8W0V0) Type IIB calcium ATPase, p... 42 4e-04
TC18250 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partia... 40 0.001
>TC10412 homologue to UP|Q9FVE8 (Q9FVE8) Plasma membrane Ca2+-ATPase,
partial (33%)
Length = 1061
Score = 313 bits (802), Expect(2) = e-159
Identities = 156/189 (82%), Positives = 171/189 (89%)
Frame = +2
Query: 539 DPQKERQACKLVKVEPFNSQKKRMGVVVELPEGGLRAHCKGASEIVLAACDNVIDSKGDV 598
D Q ERQAC LVKVEPFNS KKRM V VELP GGLRAHCKGASEIVLAACD V++S G+V
Sbjct: 2 DFQGERQACNLVKVEPFNSTKKRMSVAVELPGGGLRAHCKGASEIVLAACDKVLNSNGEV 181
Query: 599 VPLNAESRNYLESTIDQFAGEALRTLCLAYIELEHGFSAEDPIPASGYTCIGVVGIKDPV 658
VPL+ ES N+L STI+QFA EALRTLCLAY+ELE+GFSAEDPIP SG+TCIGVVGIKDPV
Sbjct: 182 VPLDEESINHLNSTINQFASEALRTLCLAYMELENGFSAEDPIPLSGFTCIGVVGIKDPV 361
Query: 659 RPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTEDGLAIEGPDFREKTQEEMF 718
RPGVKESV VCRSAGI VRMVTGDNINTAKAIARECGILT+DG+AIEGP+FREK+ EE+
Sbjct: 362 RPGVKESVAVCRSAGITVRMVTGDNINTAKAIARECGILTDDGIAIEGPEFREKSLEELL 541
Query: 719 ELIPKIQVM 727
ELIPKIQV+
Sbjct: 542 ELIPKIQVL 568
Score = 265 bits (678), Expect(2) = e-159
Identities = 134/144 (93%), Positives = 142/144 (98%)
Frame = +3
Query: 741 QLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVA 800
QLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVA
Sbjct: 630 QLRTTFGEVVAVTGDGTNDAPALHEADIGLAMGIAGTEVAKESADVIILDDNFSTIVTVA 809
Query: 801 KWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALAL 860
KWGRSVYINIQKFVQFQLTVNVVAL+VNF+SA LTG+APLTAVQLLWVNMIMDTLGALAL
Sbjct: 810 KWGRSVYINIQKFVQFQLTVNVVALIVNFTSACLTGTAPLTAVQLLWVNMIMDTLGALAL 989
Query: 861 ATEPPTDDLMKRAPLGRKGDFINS 884
ATEPP DDLMKR+P+GRKG+FI++
Sbjct: 990 ATEPPKDDLMKRSPVGRKGNFISN 1061
>BP066255
Length = 540
Score = 297 bits (760), Expect(2) = 3e-81
Identities = 144/166 (86%), Positives = 160/166 (95%)
Frame = +2
Query: 317 MLVTTVGMRTQWGKLMATLSEGGDDETPLQVKLNGVATLIGKVGLFFAVVTFVVLVKGLM 376
ML+TTVGMRTQWGKLMATL+EGGDDETPLQVKLNGVAT+IGK+GLFFA+VTF VLV+GL+
Sbjct: 38 MLITTVGMRTQWGKLMATLTEGGDDETPLQVKLNGVATIIGKIGLFFAIVTFAVLVQGLV 217
Query: 377 SRKIREGRFWWWSADDAMEMLEFFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMNDKAL 436
S K+++ FW W+ DDA+EMLE+FAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMNDKAL
Sbjct: 218 SHKLQQDSFWSWTGDDALEMLEYFAIAVTIVVVAVPEGLPLAVTLSLAFAMKKMMNDKAL 397
Query: 437 VRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMSSKEVNNK 482
VRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCICM+S+EV+NK
Sbjct: 398 VRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMNSQEVSNK 535
Score = 23.5 bits (49), Expect(2) = 3e-81
Identities = 10/10 (100%), Positives = 10/10 (100%)
Frame = +3
Query: 305 LSGTKVQDGS 314
LSGTKVQDGS
Sbjct: 3 LSGTKVQDGS 32
>TC19263 similar to UP|Q8L8A0 (Q8L8A0) Type IIB calcium ATPase MCA5, partial
(17%)
Length = 536
Score = 275 bits (703), Expect = 3e-74
Identities = 133/172 (77%), Positives = 153/172 (88%)
Frame = +3
Query: 5 LNESFGGVKSKNSTEEALSKWRKVCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQEKLRV 64
LNE+FGGVKSKNS++EAL +WRK+CGVVKNPKRRFRFTANL+KR EAAAMRR+NQEKLRV
Sbjct: 21 LNENFGGVKSKNSSQEALQRWRKLCGVVKNPKRRFRFTANLNKRTEAAAMRRSNQEKLRV 200
Query: 65 AVLVSKAAFQFIQGVQPSDYLVPDDVKAAGFHICADELGSIVEGHDVKKLKFHGGVSGIA 124
AVLVSKAA QFIQG QPS+Y VP+DVKAAGF IC DELGSIVE HDVKK KFHGGV+G+A
Sbjct: 201 AVLVSKAALQFIQGSQPSEYKVPEDVKAAGFQICGDELGSIVENHDVKKFKFHGGVNGVA 380
Query: 125 EKLSTSTTKGLSGDSEARRIRQEVYGINKFAESEVRSFWIFVYEALQDMTLM 176
+KLSTS T+G+S D++ RQ +YGINKF E E +SFW+FV+EALQDMTLM
Sbjct: 381 KKLSTSVTEGISSDADILNRRQLIYGINKFTEIEAKSFWVFVWEALQDMTLM 536
>AV779952
Length = 557
Score = 250 bits (638), Expect = 9e-67
Identities = 122/170 (71%), Positives = 143/170 (83%)
Frame = +2
Query: 530 LEFGLSLGGDPQKERQACKLVKVEPFNSQKKRMGVVVELPEGGLRAHCKGASEIVLAACD 589
L GLSLGGD K RQ KLVKVEPFNS KK MGVV++LP+GG RAHCKGASEIVLAACD
Sbjct: 47 LRIGLSLGGDFIKGRQVVKLVKVEPFNSTKKSMGVVLQLPDGGFRAHCKGASEIVLAACD 226
Query: 590 NVIDSKGDVVPLNAESRNYLESTIDQFAGEALRTLCLAYIELEHGFSAEDPIPASGYTCI 649
V+DS G+VVPL+ +S N ++ TI++FA +ALRTLCL Y++++ F PIP SGYTCI
Sbjct: 227 KVVDSNGEVVPLDEDSINQVKDTIEKFADDALRTLCLTYMDIQDEF*VGSPIPTSGYTCI 406
Query: 650 GVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTE 699
G+VGIKDPVRPGV+ESV +CRSAGI VRMVTGDNI+TAKAIARECGILT+
Sbjct: 407 GIVGIKDPVRPGVRESVAICRSAGITVRMVTGDNIDTAKAIARECGILTD 556
>TC17656 similar to GB|BAA97361.1|8843813|AB023042 Ca2+-transporting
ATPase-like protein {Arabidopsis thaliana;} , partial
(25%)
Length = 821
Score = 213 bits (542), Expect = 1e-55
Identities = 104/180 (57%), Positives = 138/180 (75%), Gaps = 3/180 (1%)
Frame = +2
Query: 801 KWGRSVYINIQKFVQFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALAL 860
+WGRSVY NIQKF+QFQLTVNV AL++N +A+ +G PL AVQLLWVN+IMDTLGALAL
Sbjct: 281 RWGRSVYANIQKFIQFQLTVNVAALVINVVAAISSGDVPLNAVQLLWVNLIMDTLGALAL 460
Query: 861 ATEPPTDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFL---RGP 917
ATEPPTD LM R+P+GR+ I +IMWRN+L QA+YQ V+ L G+ + L +
Sbjct: 461 ATEPPTDHLMDRSPVGRREPLITNIMWRNLLIQAMYQVSVLLVLNFRGRSILGLTHDKFD 640
Query: 918 NAGVVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEY 977
+A V NTLIFN+FV CQ+FNE N+R+ +E ++FKG+ N++F+ ++G TVV QI+IVE+
Sbjct: 641 HAVKVKNTLIFNAFVLCQIFNEFNARKPDEFNIFKGVTKNYLFMGIVGLTVVLQIVIVEF 820
Score = 119 bits (297), Expect = 3e-27
Identities = 66/108 (61%), Positives = 78/108 (72%), Gaps = 3/108 (2%)
Frame = +3
Query: 709 FREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADI 768
FR + E E+ I VM RSSP DK LV+ LR G VVAVTGDGTNDAPALHEADI
Sbjct: 9 FRAMSDAERDEIADAISVMGRSSPNDKLLLVQALRRK-GHVVAVTGDGTNDAPALHEADI 185
Query: 769 GLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWG---RSVYINIQKF 813
GLAMGIAGTEVAKES+D+IILDDNF+++V V+ G ++ N+ F
Sbjct: 186 GLAMGIAGTEVAKESSDIIILDDNFASVVKVSDGGGLYMQIFRNLSSF 329
>BF177756
Length = 455
Score = 204 bits (518), Expect = 7e-53
Identities = 99/150 (66%), Positives = 124/150 (82%)
Frame = +1
Query: 815 QFQLTVNVVALLVNFSSAVLTGSAPLTAVQLLWVNMIMDTLGALALATEPPTDDLMKRAP 874
QFQLTVNVVAL++NF SA +TGSAPLTAVQLLWVN+IMDTLGALALATEPP D L+KR P
Sbjct: 1 QFQLTVNVVALVINFFSACITGSAPLTAVQLLWVNLIMDTLGALALATEPPNDGLLKRPP 180
Query: 875 LGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLRGPNAGVVLNTLIFNSFVFC 934
+ R FI MWRNI+GQ++YQ +V+ L GK + L G +A VLNTLIFNSFVFC
Sbjct: 181 VARGASFITKAMWRNIIGQSIYQLIVLVILTFDGKRLLRLSGSDATRVLNTLIFNSFVFC 360
Query: 935 QVFNEINSREMEEVDVFKGIWDNHVFVAVI 964
QVFNEINSR++E++++F+G++D+ +FVA+I
Sbjct: 361 QVFNEINSRDIEKINIFRGMFDSWIFVAII 450
>AV409404
Length = 421
Score = 166 bits (419), Expect = 2e-41
Identities = 86/139 (61%), Positives = 103/139 (73%), Gaps = 2/139 (1%)
Frame = +1
Query: 421 LSLAFAMKKMMNDKALVRHLAACETMGSATTICSDKTGTLTTNHMTVVKTCICMSSKEVN 480
LSLAFAMKK+MND+ALVRHL+ACETMGSA IC+DKTGTLTTNHM V K IC + E+
Sbjct: 4 LSLAFAMKKLMNDRALVRHLSACETMGSANCICTDKTGTLTTNHMVVDKIWICEKTTEIK 183
Query: 481 NKE--HGLCSELPDSAQKLLLQSIFNNTGGEVVVNKRGKREILGTPTESAILEFGLSLGG 538
E L SE+ + + LQ+IF NT EVV +K GK+ ILGTPTESA+LEFGL GG
Sbjct: 184 GNESVEKLKSEISEEVISIFLQAIFQNTSSEVVNDKEGKKAILGTPTESALLEFGLLSGG 363
Query: 539 DPQKERQACKLVKVEPFNS 557
D +R+ K++KVEPFNS
Sbjct: 364 DFDAQRRDYKILKVEPFNS 420
>AV417225
Length = 359
Score = 144 bits (362), Expect = 9e-35
Identities = 69/119 (57%), Positives = 90/119 (74%)
Frame = +3
Query: 856 GALALATEPPTDDLMKRAPLGRKGDFINSIMWRNILGQALYQFVVIWFLQTVGKWVFFLR 915
GALALATEPP D LM+R P+GR+ FI MWRNI GQ++YQ +V+ L GK + L
Sbjct: 3 GALALATEPPNDGLMERLPVGRRASFITKPMWRNIFGQSIYQLIVLGVLNFDGKRLLGLT 182
Query: 916 GPNAGVVLNTLIFNSFVFCQVFNEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIII 974
G +A VLNT+IFN+FVFCQVFNEINSRE+E++++F+G++D+ +F VI TV FQ II
Sbjct: 183 GSDATAVLNTVIFNTFVFCQVFNEINSREIEKINIFRGMFDSGIFFTVIFSTVAFQAII 359
>AV780873
Length = 487
Score = 127 bits (318), Expect = 1e-29
Identities = 64/148 (43%), Positives = 97/148 (65%)
Frame = +3
Query: 100 DELGSIVEGHDVKKLKFHGGVSGIAEKLSTSTTKGLSGDSEARRIRQEVYGINKFAESEV 159
++L SI HD L+ +GGV+ I+ L T+ KG+ D R+ +G N + +
Sbjct: 36 EQLASISREHDTAALQQYGGVAVISNLLKTNLEKGVIDDDADLLKRKNAFGSNNYPRKKG 215
Query: 160 RSFWIFVYEALQDMTLMILAVCAFVSLIVGIATEGWPQGSHDGLGIVASILLVVFVTATS 219
R FW+F+++A +D+TL+IL V A SL +GI +EG +G +DG I ++LLV+FVTA S
Sbjct: 216 RPFWMFLWDACKDLTLIILMVAAAASLALGIKSEGIEEGWYDGGSIAFAVLLVIFVTAVS 395
Query: 220 DYRQSLQFKDLDKEKKKISIQVTRNGYR 247
DY+QSLQF+DL++EK+ I ++V R G R
Sbjct: 396 DYKQSLQFRDLNEEKRNIHLEVIRGGRR 479
>AV777297
Length = 610
Score = 113 bits (283), Expect = 1e-25
Identities = 64/156 (41%), Positives = 94/156 (60%)
Frame = +1
Query: 1 MESYLNESFGGVKSKNSTEEALSKWRKVCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQE 60
MES L + +++K+ + E LSKWR +VKNP+RRFR A+L KR +A + Q
Sbjct: 139 MESLLKDF--ELENKDHSIEGLSKWRSAVSLVKNPRRRFRNVADLVKRLQAQEKLKKIQG 312
Query: 61 KLRVAVLVSKAAFQFIQGVQPSDYLVPDDVKAAGFHICADELGSIVEGHDVKKLKFHGGV 120
+R + +AA QF + V ++Y + + + AGF I D++ SIV GHD K L+ G V
Sbjct: 313 TIRAVIFAQRAALQFKEAVGATEYKLSEKTREAGFGIEPDDIASIVRGHDHKNLRKVGKV 492
Query: 121 SGIAEKLSTSTTKGLSGDSEARRIRQEVYGINKFAE 156
GIA KLS S +G+S + RQEVYG+N+++E
Sbjct: 493 EGIASKLSVSIDEGVS--QHSINSRQEVYGVNRYSE 594
>BP064305
Length = 510
Score = 112 bits (280), Expect = 3e-25
Identities = 62/140 (44%), Positives = 89/140 (63%), Gaps = 1/140 (0%)
Frame = +1
Query: 12 VKSKNSTEEALSKWRK-VCGVVKNPKRRFRFTANLSKRYEAAAMRRTNQEKLRVAVLVSK 70
++ KN + EAL +WR V VVKN +RRFR A+L KR EA +++ +EK+R+A+ V K
Sbjct: 97 LEHKNPSVEALRRWRSAVSFVVKNRRRRFRMCADLDKRSEAEQIKQGIKEKIRIALYVQK 276
Query: 71 AAFQFIQGVQPSDYLVPDDVKAAGFHICADELGSIVEGHDVKKLKFHGGVSGIAEKLSTS 130
AA QFI +Y +P++ + AGF I ADE+ S+V HD K L +GGV +A KLS S
Sbjct: 277 AALQFIDAGNRVEYELPEEAREAGFGIHADEVASLVRSHDYKNLSNNGGVEALARKLSVS 456
Query: 131 TTKGLSGDSEARRIRQEVYG 150
+G+S + RQ++YG
Sbjct: 457 VDEGVS--EASINSRQQIYG 510
>TC17282 similar to UP|Q8L8A0 (Q8L8A0) Type IIB calcium ATPase MCA5, partial
(7%)
Length = 498
Score = 108 bits (271), Expect = 3e-24
Identities = 50/73 (68%), Positives = 62/73 (84%)
Frame = +1
Query: 942 SREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLSVGYV 1001
SREME+++V GI +N+VFVAV+ T +FQIIIVEY+GTFANTTPL+LVQW FCL VG++
Sbjct: 1 SREMEKINVLNGILENYVFVAVLSATALFQIIIVEYMGTFANTTPLTLVQWFFCLVVGFL 180
Query: 1002 GMPIATYLKQIPV 1014
GMPIA +K IPV
Sbjct: 181 GMPIAAGIKMIPV 219
>TC11343 similar to PIR|B96660|B96660 protein F2K11.18 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(21%)
Length = 909
Score = 100 bits (249), Expect = 1e-21
Identities = 61/177 (34%), Positives = 95/177 (53%)
Frame = +2
Query: 649 IGVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTEDGLAIEGPD 708
IGV+ + DP++P +E V + S I MVTGDN TA +IAR+ GI T
Sbjct: 164 IGVLAVSDPLKPNAREVVSILNSMNIRSIMVTGDNWGTANSIARQAGIET---------- 313
Query: 709 FREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADI 768
V+A + P K T VK+L+T+ G VA+ GDG ND+PAL AD+
Sbjct: 314 -----------------VIAEAQPQTKATKVKELQTS-GYTVAMVGDGINDSPALVAADV 439
Query: 769 GLAMGIAGTEVAKESADVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVAL 825
G+A+G AGT++A E++D++++ N I+ + + I+ + L N++A+
Sbjct: 440 GIAIG-AGTDIAIEASDIVLMRSNLEDIIIALDLAKKTFSRIRLNYLWALGYNLLAI 607
>TC14980 similar to UP|ACA9_ARATH (Q9LU41) Potential calcium-transporting
ATPase 9, plasma membrane-type (Ca(2+)-ATPase isoform 9)
, partial (8%)
Length = 594
Score = 69.3 bits (168), Expect = 3e-12
Identities = 31/77 (40%), Positives = 50/77 (64%)
Frame = +1
Query: 938 NEINSREMEEVDVFKGIWDNHVFVAVIGCTVVFQIIIVEYLGTFANTTPLSLVQWIFCLS 997
NE N+R+ EE++VF+G+ N +F+ ++ T + QIII+E+LG F +T L+ W+ L
Sbjct: 1 NEFNARKPEEMNVFRGVTKNRLFMGIVVMTFILQIIIIEFLGKFTDTVRLNWTLWLASLL 180
Query: 998 VGYVGMPIATYLKQIPV 1014
+G + P+A K IPV
Sbjct: 181 IGLISWPLAIAGKFIPV 231
>TC19919 homologue to UP|Q7Y068 (Q7Y068) Plasma membrane H+-ATPase, partial
(21%)
Length = 586
Score = 63.9 bits (154), Expect = 1e-10
Identities = 63/238 (26%), Positives = 100/238 (41%)
Frame = +1
Query: 442 ACETMGSATTICSDKTGTLTTNHMTVVKTCICMSSKEVNNKEHGLCSELPDSAQKLLLQS 501
A E M +CSDKTGTLT N ++V + I + +K + KEH
Sbjct: 1 AIEEMAGMDVLCSDKTGTLTLNKLSVDRNLIEVFTKGIE-KEH----------------- 126
Query: 502 IFNNTGGEVVVNKRGKREILGTPTESAILEFGLSLGGDPQKERQACKLVKVEPFNSQKKR 561
+++ R R T + AI + + DP++ R + V PFN KR
Sbjct: 127 -------VILLAARASR----TENQDAIDAAIVGMLADPKEARAGVREVHFLPFNPVDKR 273
Query: 562 MGVVVELPEGGLRAHCKGASEIVLAACDNVIDSKGDVVPLNAESRNYLESTIDQFAGEAL 621
+ +G KGA E +L N+ + K DV R STID+FA L
Sbjct: 274 TALTYIDSDGSWHRASKGAPEQIL----NLCNCKEDV-------RKRAHSTIDKFAERGL 420
Query: 622 RTLCLAYIELEHGFSAEDPIPASGYTCIGVVGIKDPVRPGVKESVQVCRSAGIMVRMV 679
R+L +A + P + + +G++ + DP R E++ + G+ V+M+
Sbjct: 421 RSLGVARQVVP---EKSKDAPGAPWQFVGLLPLFDPPRHDSAETITRALNLGVNVKMI 585
>AV781319
Length = 524
Score = 59.3 bits (142), Expect = 3e-09
Identities = 28/67 (41%), Positives = 45/67 (66%)
Frame = +1
Query: 150 GINKFAESEVRSFWIFVYEALQDMTLMILAVCAFVSLIVGIATEGWPQGSHDGLGIVASI 209
G N + + RSF +F+++A +D+TL+IL V A SL +GI +EG +G +DG I ++
Sbjct: 118 GSNNYPRKKGRSFLMFIWDAGKDLTLIILTVAAIASLALGIKSEGIKEGWYDGGSIAVAV 297
Query: 210 LLVVFVT 216
+LV+ VT
Sbjct: 298 ILVILVT 318
>TC10275 similar to UP|Q7Y051 (Q7Y051) Paa2 P-type ATPase, partial (16%)
Length = 724
Score = 55.1 bits (131), Expect = 6e-08
Identities = 32/101 (31%), Positives = 53/101 (51%), Gaps = 1/101 (0%)
Frame = +1
Query: 726 VMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALHEADIGLAM-GIAGTEVAKESA 784
V A +P K + L+ G VA+ GDG NDAPAL AD+G+A+ A A ++A
Sbjct: 7 VKASLAPQQKSEFISSLKAA-GHHVAMVGDGINDAPALAVADVGIALQNEAQENAASDAA 183
Query: 785 DVIILDDNFSTIVTVAKWGRSVYINIQKFVQFQLTVNVVAL 825
+I+L + S +V +S + + + + + NV+A+
Sbjct: 184 SIILLGNKISQVVDAIDLAQSTMAKVYQNLSWAVAYNVIAI 306
>AU088926
Length = 487
Score = 44.3 bits (103), Expect = 1e-04
Identities = 36/126 (28%), Positives = 56/126 (43%), Gaps = 4/126 (3%)
Frame = +2
Query: 649 IGVVGIKDPVRPGVKESVQVCRSAGIMVRMVTGDNINTAKAIARECGILTE----DGLAI 704
+G++ + DP R +++ G+ V+M+TGD + K R G+ T L
Sbjct: 20 LGLLPLFDPPRHDSAXTIRRALDLGVNVKMITGDQLAIGKETGRRLGMGTNMXPSSSLLG 199
Query: 705 EGPDFREKTQEEMFELIPKIQVMARSSPLDKHTLVKQLRTTFGEVVAVTGDGTNDAPALH 764
+ D + ELI K A P K +V +L T + +TG G NDAPAL
Sbjct: 200 QSKD-AAIASIPIDELIEKADGFAGVFPEHKXEIVXRLXDT-KHICGMTGXGVNDAPALX 373
Query: 765 EADIGL 770
+ + L
Sbjct: 374 KXTLVL 391
>TC18638 similar to UP|Q8W0V0 (Q8W0V0) Type IIB calcium ATPase, partial (6%)
Length = 464
Score = 42.4 bits (98), Expect = 4e-04
Identities = 23/45 (51%), Positives = 28/45 (62%)
Frame = +3
Query: 970 FQIIIVEYLGTFANTTPLSLVQWIFCLSVGYVGMPIATYLKQIPV 1014
FQ IIV++LGTFAN PL+ + + G V MPI LK IPV
Sbjct: 18 FQPIIVKFLGTFANPVPLNWQLRLLSVLFGAVSMPIFAILKCIPV 152
>TC18250 similar to UP|Q8L741 (Q8L741) AT3g13690/MMM17_12, partial (3%)
Length = 757
Score = 40.4 bits (93), Expect = 0.001
Identities = 19/29 (65%), Positives = 22/29 (75%)
Frame = +1
Query: 295 VMVTSQNPFLLSGTKVQDGSCTMLVTTVG 323
++ TSQN F L GTKVQDGS TMLV +G
Sbjct: 313 ILQTSQN*FFLPGTKVQDGSSTMLVILIG 399
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,016,136
Number of Sequences: 28460
Number of extensions: 185559
Number of successful extensions: 992
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 979
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 986
length of query: 1014
length of database: 4,897,600
effective HSP length: 99
effective length of query: 915
effective length of database: 2,080,060
effective search space: 1903254900
effective search space used: 1903254900
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0347b.6


