

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0343.3
(521 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
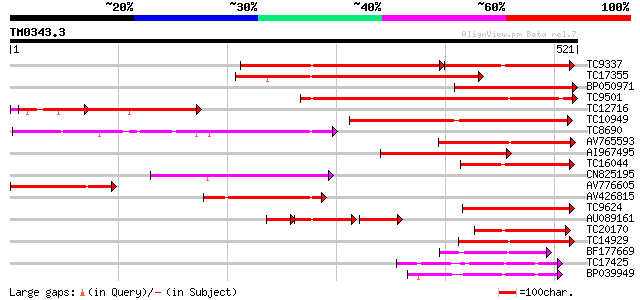
Score E
Sequences producing significant alignments: (bits) Value
TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B pre... 285 e-123
TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%) 236 5e-63
BP050971 226 5e-60
TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesteras... 224 3e-59
TC12716 weakly similar to PIR|T49241|T49241 pectinesterase-like ... 217 4e-57
TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precu... 213 8e-56
TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precu... 159 1e-39
AV765593 134 5e-32
AI967495 125 2e-29
TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase ... 124 4e-29
CN825195 120 4e-28
AV776605 108 2e-24
AV426815 107 4e-24
TC9624 similar to UP|Q43234 (Q43234) Pectinmethylesterase precur... 107 4e-24
AU089161 62 9e-24
TC20170 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isof... 106 1e-23
TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase ... 106 1e-23
BF177669 101 3e-22
TC17425 weakly similar to UP|Q9FM79 (Q9FM79) Similarity to pecti... 100 1e-21
BP039949 91 5e-19
>TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (54%)
Length = 1145
Score = 285 bits (728), Expect(2) = e-123
Identities = 140/187 (74%), Positives = 159/187 (84%)
Frame = +2
Query: 213 DGSGNYTTVMDAVKDAPEYNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISG 272
DGSGNYTTVM+AV AP+Y+M R+VI+VKKG+Y+EY EIKKKKWNIM+VGDGMDVT ISG
Sbjct: 29 DGSGNYTTVMEAVLAAPDYSMKRFVIYVKKGVYEEYAEIKKKKWNIMMVGDGMDVTVISG 208
Query: 273 NRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFYRCGIK 332
NRS+G D TTF++ATFAVS GFIAR ITF+NTAGPE QAVALRSDSDL+VF+RC I
Sbjct: 209 NRSFG-DGWTTFKSATFAVSGRGFIARGITFQNTAGPEKHQAVALRSDSDLAVFFRCSIT 385
Query: 333 GYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTH 392
GYQDSLY H+ RQFYKECRI GTVDFI G+A AVFQNC IQ +K L+DQKNTI AQ R
Sbjct: 386 GYQDSLYPHTMRQFYKECRITGTVDFIFGDATAVFQNCNIQARKGLQDQKNTITAQGRKD 565
Query: 393 PNQTSGF 399
P Q+SGF
Sbjct: 566 PGQSSGF 586
Score = 176 bits (445), Expect(2) = e-123
Identities = 80/120 (66%), Positives = 97/120 (80%)
Frame = +3
Query: 400 SFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYD 459
SF C I D + PS GSIQT+LGRPWK +SRTVFM+SFMS+++ +GWLEW NT++D
Sbjct: 588 SFQYCNITADSELLPSVGSIQTYLGRPWKEYSRTVFMQSFMSDVVSSQGWLEW--NTSFD 761
Query: 460 DTLFFGEYQNKGPGSGVANRVKWRGYHVLNNSQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
DTLF+GEY N G G+GV NRVKW GYHVLN+S+A FTVNQFI+G+LWLPSTGV+Y GL
Sbjct: 762 DTLFYGEYLNYGAGAGVGNRVKWGGYHVLNDSEAGKFTVNQFIEGNLWLPSTGVAYTGGL 941
>TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%)
Length = 848
Score = 236 bits (603), Expect = 5e-63
Identities = 119/231 (51%), Positives = 157/231 (67%), Gaps = 3/231 (1%)
Frame = +1
Query: 208 VVVAADGSGNYTTVMDAVKDAPEYNMTR---YVIHVKKGIYKEYVEIKKKKWNIMIVGDG 264
+V +G+ NYT++ DA+ AP + + ++I+V++G Y+EYV + K+K NI++VGDG
Sbjct: 157 IVSPYNGTDNYTSIGDAIAAAPNHTKPQDGYFLIYVREGYYEEYVIVPKEKKNILLVGDG 336
Query: 265 MDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLS 324
++ T I+GN S D TTF ++TFAVS F A D+TF NTAGP QAVA+R+++DLS
Sbjct: 337 INKTIITGNHSV-IDGWTTFNSSTFAVSGERFTAVDVTFRNTAGPAKHQAVAVRNNADLS 513
Query: 325 VFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNT 384
FYRC +GYQD+LY HS RQFY+EC I GTVDFI GNAA VFQNC I +KPL QKN
Sbjct: 514 TFYRCSFEGYQDTLYVHSLRQFYRECDIYGTVDFIFGNAAVVFQNCNIYARKPLPFQKNA 693
Query: 385 IAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVF 435
+ AQ RT PNQ +G S NCTI PD S ++LGRPW+ + F
Sbjct: 694 VTAQGRTDPNQNTGISIQNCTIDAAPDLAAELNSTLSYLGRPWEGLFKDWF 846
>BP050971
Length = 477
Score = 226 bits (577), Expect = 5e-60
Identities = 105/113 (92%), Positives = 106/113 (92%)
Frame = -1
Query: 409 DPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQ 468
DPDFQPSNGSI TFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWET TTYDD LFFGEY
Sbjct: 477 DPDFQPSNGSIPTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETYTTYDDPLFFGEYP 298
Query: 469 NKGPGSGVANRVKWRGYHVLNNSQAMDFTVNQFIKGDLWLPSTGVSYNAGLLV 521
NKGPGSGVANRVKWRGYHVLNNS AMDFTVN FIKGDLWLPS GVSY+AGLLV
Sbjct: 297 NKGPGSGVANRVKWRGYHVLNNSPAMDFTVNPFIKGDLWLPSPGVSYHAGLLV 139
>TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase ,
partial (29%)
Length = 934
Score = 224 bits (570), Expect = 3e-59
Identities = 112/255 (43%), Positives = 157/255 (60%), Gaps = 1/255 (0%)
Frame = +2
Query: 268 TTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALRSDSDLSVFY 327
T I+G +++ D + T +TATFA A GFIA+ ITFENTAGP QAVALR+ D S F
Sbjct: 14 TIITGRKNFA-DGVKTMQTATFANQAPGFIAKAITFENTAGPNGHQAVALRNQGDQSAFV 190
Query: 328 RCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAA 387
C I GYQD+LY + RQFY+ C I GT+DFI G + + Q+ +I +KP +Q NT+ A
Sbjct: 191 GCHILGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSPTLIQHSIIVVRKPNDNQFNTVTA 370
Query: 388 QSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPE 447
N +G +C I+P+ P I+++LGRPWK +SRTV MES + + IHPE
Sbjct: 371 DGTPTKNMPTGIVIQDCEIVPEAALFPVRFKIKSYLGRPWKEYSRTVVMESTLGDFIHPE 550
Query: 448 GWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYH-VLNNSQAMDFTVNQFIKGDL 506
GW W + DTL++ E+ N GPG+ A R+KW+GY V++ ++A FT +F+K
Sbjct: 551 GWFPWAGSFAL-DTLYYAEHANTGPGANTAGRIKWKGYRGVISRAEATQFTAGEFLKAG- 724
Query: 507 WLPSTGVSYNAGLLV 521
P +G+ + L V
Sbjct: 725 --PRSGLDWIKALHV 763
>TC12716 weakly similar to PIR|T49241|T49241 pectinesterase-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(11%)
Length = 709
Score = 217 bits (552), Expect = 4e-57
Identities = 124/179 (69%), Positives = 137/179 (76%), Gaps = 11/179 (6%)
Frame = -3
Query: 9 PTLILLLIMSLQPCTTPSSASTITEFSGLSCLRVS------PNRFIDSANKVINLLQ-NV 61
P +LLIMSLQPCTTP + F LR+S PNR S++K + +
Sbjct: 524 PHSSVLLIMSLQPCTTP-----VFRFYHH*RLRISHVSELSPNRVHWSSDKQSH*SSCRM 360
Query: 62 SYNLS-PFTKVNQDNDNRLSNAISDCLELFDMAYENLKWSISATQNHQ---GNKNNGTGN 117
S+ +S P K+ +DNDNRLSNAISDCLE+ DMAYENL WSISATQNHQ GNKNNGTGN
Sbjct: 359 SHTISHPSPKLIRDNDNRLSNAISDCLEVLDMAYENLTWSISATQNHQDIPGNKNNGTGN 180
Query: 118 LSSDLRTWISAVLSYQETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLLQVVPDSDP 176
LSSDLRTWISAVLSYQETC+DGFEGTNS VKDHVSGGLDQVISLVKNELLLQVVPDSDP
Sbjct: 179 LSSDLRTWISAVLSYQETCMDGFEGTNSKVKDHVSGGLDQVISLVKNELLLQVVPDSDP 3
Score = 48.1 bits (113), Expect = 3e-06
Identities = 37/83 (44%), Positives = 41/83 (48%), Gaps = 10/83 (12%)
Frame = -2
Query: 1 MAISSTLTPTLILLL----------IMSLQPCTTPSSASTITEFSGLSCLRVSPNRFIDS 50
MAISSTLTPTLI L + L P S + FS S L +
Sbjct: 549 MAISSTLTPTLIRLADNVSSTMHHPSLPLLPSLKAQD*SCLRAFSKQSSLEL*QT----- 385
Query: 51 ANKVINLLQNVSYNLSPFTKVNQ 73
K + LQNVSYNLSPFTKVNQ
Sbjct: 384 --KSLIFLQNVSYNLSPFTKVNQ 322
>TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (34%)
Length = 773
Score = 213 bits (541), Expect = 8e-56
Identities = 103/206 (50%), Positives = 133/206 (64%), Gaps = 1/206 (0%)
Frame = +3
Query: 313 QAVALRSDSDLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICGNAAAVFQNCLI 372
QAVA+RS SD SVFYRC G+QD+LYAHS RQFY+EC I GT+DFI GNAA++FQNC I
Sbjct: 12 QAVAMRSGSDRSVFYRCSFDGFQDTLYAHSNRQFYRECDITGTIDFIFGNAASIFQNCKI 191
Query: 373 QPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSR 432
P++P+ +Q TI AQ + PNQ +G TI P S + T+LGRPWK+FS
Sbjct: 192 MPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITP----LESTYTAPTYLGRPWKDFST 359
Query: 433 TVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGY-HVLNNS 491
T+ M+S + + P GW+ W N T+F+ E+QN GPGS VA RVKW GY L
Sbjct: 360 TLIMQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLTAD 539
Query: 492 QAMDFTVNQFIKGDLWLPSTGVSYNA 517
FT+ FI+G WLP T V++++
Sbjct: 540 DLGKFTLASFIQGPEWLPDTAVAFDS 617
>TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor
(Pectinesterase) , partial (59%)
Length = 1082
Score = 159 bits (401), Expect = 1e-39
Identities = 112/311 (36%), Positives = 164/311 (52%), Gaps = 12/311 (3%)
Frame = +3
Query: 3 ISSTLTPTLILLLIMSLQPCTTPSSAST-ITEFSGLSCLRVSPNRFIDSANKVINLLQNV 61
I+S L PT L + C AS+ + S +S +S + N +I+L+
Sbjct: 186 IASYLKPTSFNLFLSPPHGCEHALDASSCLAHVSEVSQSPISATKD-PKLNILISLMTKS 362
Query: 62 SYNLSPFTKVNQDNDNRLSN-----AISDCLELFDMAYENLKWSISATQNHQGNKNNGTG 116
+ ++ + NR++N A+SDC +L D++ + + S+ A K+N
Sbjct: 363 TSHIQEAMVKTKAIKNRINNPREEAALSDCEQLMDLSIDRVWDSVMAL-----TKDNTDS 527
Query: 117 NLSSDLRTWISAVLSYQETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLLQV---VPD 173
+ D W+S VL+ TC+DG EG + + + ++ +IS K L L V P
Sbjct: 528 H--QDAHAWLSGVLTNHATCLDGLEGPSRALME---AEIEDLISRSKTSLALLVSVLAPK 692
Query: 174 SDPSNISNS---GEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPE 230
I + G+FPSWV +D +LL+ A+VVVA DGSG + TV +AV AP+
Sbjct: 693 GGNEQIIDEPLDGDFPSWVTRKDRRLLESSVGDVNANVVVAKDGSGRFKTVAEAVASAPD 872
Query: 231 YNMTRYVIHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFA 290
TRYVI+VKKG YKE +EI KKK N+M+ GDGMD T I+GN + D TTF++AT A
Sbjct: 873 SGKTRYVIYVKKGTYKENIEIGKKKTNVMLTGDGMDATIITGNLNV-IDGSTTFKSATVA 1049
Query: 291 VSAIGFIARDI 301
GFIA+DI
Sbjct: 1050AVGDGFIAQDI 1082
>AV765593
Length = 501
Score = 134 bits (336), Expect = 5e-32
Identities = 61/126 (48%), Positives = 80/126 (63%)
Frame = -1
Query: 395 QTSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWET 454
Q +G S HNCTI D SNG+ T+LGRPWK +SRTV+M++ M +I GW W+
Sbjct: 501 QDTGTSIHNCTIRAADDLASSNGATATYLGRPWKEYSRTVYMQTSMDNVIDSAGWRAWDG 322
Query: 455 NTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNSQAMDFTVNQFIKGDLWLPSTGVS 514
TL++ E+ N GPGS RV W+GYHV+N + A +FTV+ F+ GD WLP TGVS
Sbjct: 321 EFALS-TLYYAEFNNSGPGSNTNGRVTWQGYHVINATDAANFTVSNFLLGDDWLPQTGVS 145
Query: 515 YNAGLL 520
Y L+
Sbjct: 144 YTNTLI 127
>AI967495
Length = 390
Score = 125 bits (314), Expect = 2e-29
Identities = 60/121 (49%), Positives = 74/121 (60%)
Frame = +1
Query: 341 HSQRQFYKECRIRGTVDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFS 400
H+QRQFYK+C+I GTVD I GNAA VFQNC I KKPL Q N I AQ R P Q +G +
Sbjct: 19 HAQRQFYKQCQIYGTVDLIFGNAAVVFQNCKIFAKKPLDGQANMITAQGRDDPFQNTGIT 198
Query: 401 FHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDD 460
HN I PD +P T LGRPW+ +SR V +++FM ++ P GW W+ D
Sbjct: 199 IHNSEIRAAPDLKPVVNKHNTSLGRPWQQYSRVVVLKTFMDTLVSPLGWSSWDDTDFALD 378
Query: 461 T 461
T
Sbjct: 379 T 381
>TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase (Fragment)
, partial (43%)
Length = 564
Score = 124 bits (311), Expect = 4e-29
Identities = 59/107 (55%), Positives = 78/107 (72%), Gaps = 2/107 (1%)
Frame = +3
Query: 415 SNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYD-DTLFFGEYQNKGPG 473
SN S +T+LGRPWK +SRTV+M S++ + +H GWLEW NTT+ DTL++GEY N GPG
Sbjct: 12 SNDSFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEW--NTTFALDTLYYGEYMNYGPG 185
Query: 474 SGVANRVKWRGYHVLNNS-QAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
+ V RV W GY V+ ++ +A FTV QFI G WLPSTGV++ +GL
Sbjct: 186 AAVGQRVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGL 326
>CN825195
Length = 551
Score = 120 bits (302), Expect = 4e-28
Identities = 66/178 (37%), Positives = 99/178 (55%), Gaps = 10/178 (5%)
Frame = +2
Query: 130 LSYQETCIDGFEGTNSNVKDHVSGGLDQVISLVKNELLLQVVPDSDPSNIS--------- 180
L+ Q+TC DGF T+ +VKD ++ L + LV N L + D S +
Sbjct: 17 LTNQDTCADGFADTSGDVKDQMAVNLKDLSELVSNCLAIFAGAGDDFSGVPIQNRRLLAM 196
Query: 181 NSGEFPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYVIHV 240
FP W+ D +LL ADVVV+ DG+G T+ +A+K PEY R++I+V
Sbjct: 197 RDDNFPRWLNRRDRRLLSLPVSAMQADVVVSKDGNGTAKTIAEAIKKVPEYGSRRFIIYV 376
Query: 241 KKGIYKE-YVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFI 297
+ G Y+E ++I +K+ N+MI+GDG T I+G ++ + +TTF TA+FA S GFI
Sbjct: 377 RAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMYGLLTTFHTASFAASGAGFI 550
>AV776605
Length = 323
Score = 108 bits (271), Expect = 2e-24
Identities = 60/99 (60%), Positives = 78/99 (78%), Gaps = 1/99 (1%)
Frame = +1
Query: 1 MAISSTLTPT-LILLLIMSLQPCTTPSSASTITEFSGLSCLRVSPNRFIDSANKVINLLQ 59
M +SSTLTPT LILLLI+SLQ CTT SSA+++T+FSG +CLRVS RFIDS +KVI++L
Sbjct: 28 MVLSSTLTPTTLILLLILSLQACTTQSSAASVTDFSGSACLRVSTTRFIDSVSKVIDVLG 207
Query: 60 NVSYNLSPFTKVNQDNDNRLSNAISDCLELFDMAYENLK 98
+VS L PF ++ RL+NA+SDCL+L DMA E+L+
Sbjct: 208 DVSSILKPFAS-RIFHNFRLTNAVSDCLDLIDMASEDLR 321
>AV426815
Length = 391
Score = 107 bits (268), Expect = 4e-24
Identities = 61/114 (53%), Positives = 77/114 (67%), Gaps = 1/114 (0%)
Frame = +3
Query: 179 ISNSGE-FPSWVKAEDEKLLQQDAPPPPADVVVAADGSGNYTTVMDAVKDAPEYNMTRYV 237
+ +GE +P W+ A D +LLQ A ADVVVAADGSGN+ TV AV APE + TRYV
Sbjct: 42 LMEAGEVWPEWISAADRRLLQ--AGTVKADVVVAADGSGNFKTVSAAVAAAPEKSKTRYV 215
Query: 238 IHVKKGIYKEYVEIKKKKWNIMIVGDGMDVTTISGNRSYGHDNITTFRTATFAV 291
I +K G+Y+E VE+ KKK NIM +GDG T I+G+R+ D TTF +AT AV
Sbjct: 216 IKIKAGVYRENVEVPKKKTNIMFLGDGRTTTIITGSRNV-VDGSTTFHSATVAV 374
>TC9624 similar to UP|Q43234 (Q43234) Pectinmethylesterase precursor
(Pectinesterase) (Pectin methylesterase) (Fragment) ,
partial (33%)
Length = 586
Score = 107 bits (268), Expect = 4e-24
Identities = 48/105 (45%), Positives = 70/105 (65%), Gaps = 2/105 (1%)
Frame = +3
Query: 417 GSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNT-TYDDTLFFGEYQNKGPGSG 475
GSI+T+LGRPWK +SRT+ ++S + I P GW EW+ + + TL++GEY N G G+G
Sbjct: 6 GSIKTYLGRPWKKYSRTIVLQSSIDSHIDPTGWAEWDAQSKDFLQTLYYGEYLNSGAGAG 185
Query: 476 VANRVKWRGYHVLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
RV W G+HV+ ++A FTV Q I+G++WL GV++ GL
Sbjct: 186 TGKRVNWPGFHVIKTAAEASKFTVAQLIQGNVWLKGKGVNFIEGL 320
>AU089161
Length = 400
Score = 62.0 bits (149), Expect(3) = 9e-24
Identities = 26/40 (65%), Positives = 30/40 (75%)
Frame = +2
Query: 322 DLSVFYRCGIKGYQDSLYAHSQRQFYKECRIRGTVDFICG 361
D S F+ C I GYQD+LYAH+ RQFY+ C I GTVDFI G
Sbjct: 263 DRSAFFDCAISGYQDTLYAHAHRQFYRNCEISGTVDFIFG 382
Score = 48.9 bits (115), Expect(3) = 9e-24
Identities = 27/57 (47%), Positives = 35/57 (61%)
Frame = +1
Query: 262 GDGMDVTTISGNRSYGHDNITTFRTATFAVSAIGFIARDITFENTAGPENKQAVALR 318
G G T I+GN+++ D T R+ATF A F+A+ + FENTAG QAVALR
Sbjct: 85 GXGPKKTVITGNKNFV-DGYKTMRSATFTTVAPDFMAKSMGFENTAGASKHQAVALR 252
Score = 36.2 bits (82), Expect(3) = 9e-24
Identities = 15/26 (57%), Positives = 21/26 (80%)
Frame = +3
Query: 237 VIHVKKGIYKEYVEIKKKKWNIMIVG 262
VI+VK G+Y EY++I KKK NI++ G
Sbjct: 9 VIYVKAGVYDEYIQIDKKKPNILMYG 86
>TC20170 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (32%)
Length = 592
Score = 106 bits (264), Expect = 1e-23
Identities = 50/89 (56%), Positives = 67/89 (75%), Gaps = 1/89 (1%)
Frame = +1
Query: 428 KNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWRGYHV 487
K +SRTVF++S+M ++I P GWLEW +T DTL++GEYQN+GPGS + RVKW GY V
Sbjct: 1 KLYSRTVFLKSYMEDLIDPAGWLEWN-DTFALDTLYYGEYQNRGPGSNPSARVKWGGYRV 177
Query: 488 LNNS-QAMDFTVNQFIKGDLWLPSTGVSY 515
+N+S +A FTV QFI+G WL STG+ +
Sbjct: 178 INSSTEASQFTVGQFIQGSDWLNSTGIPF 264
>TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase (Fragment)
, partial (24%)
Length = 693
Score = 106 bits (264), Expect = 1e-23
Identities = 52/108 (48%), Positives = 69/108 (63%), Gaps = 1/108 (0%)
Frame = +2
Query: 413 QPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGP 472
+P S T+LGRPWK +SRTVFM+S +S++I P GW EW N +TL + EYQ GP
Sbjct: 2 EPVKYSFPTYLGRPWKEYSRTVFMQSSISDVIDPAGWHEWSGNFAL-NTLVYREYQYPGP 178
Query: 473 GSGVANRVKWRGYHVLNN-SQAMDFTVNQFIKGDLWLPSTGVSYNAGL 519
G+G + RV W+G V+ + S+A FT FI G WL STG ++ GL
Sbjct: 179 GAGTSKRVAWKGVKVITSASEAQAFTPGSFIAGSNWLGSTGFPFSLGL 322
>BF177669
Length = 336
Score = 101 bits (251), Expect = 3e-22
Identities = 48/103 (46%), Positives = 62/103 (59%)
Frame = +3
Query: 396 TSGFSFHNCTIIPDPDFQPSNGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETN 455
T G S NC+I P + +QT+LGRPWKN+S TV M+S + I P GWL W N
Sbjct: 36 TQGISIQNCSISPFGNLS----YVQTYLGRPWKNYSTTVVMQSTLGSFISPNGWLPWVGN 203
Query: 456 TTYDDTLFFGEYQNKGPGSGVANRVKWRGYHVLNNSQAMDFTV 498
+ DT+F+ EYQN G G+ NRVKW+G + + QA FTV
Sbjct: 204 SA-PDTIFYAEYQNVGQGASTKNRVKWKGLKTITSKQASKFTV 329
>TC17425 weakly similar to UP|Q9FM79 (Q9FM79) Similarity to pectin
methylesterase, partial (33%)
Length = 643
Score = 99.8 bits (247), Expect = 1e-21
Identities = 56/153 (36%), Positives = 82/153 (52%)
Frame = +1
Query: 356 VDFICGNAAAVFQNCLIQPKKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPS 415
VDFICGNA ++F C++ + + IAA R P++ +GFSF +CTI
Sbjct: 1 VDFICGNAKSLFHECILYS---VAEFWGAIAAHHRESPDEDTGFSFVDCTI-------KG 150
Query: 416 NGSIQTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSG 475
NGS+ LGR W ++ T++ M ++I P GW +W+ + T FGEYQ G GS
Sbjct: 151 NGSV--LLGRAWGEYATTIYSNCDMDDIISPMGWSDWDV-PSRQRTALFGEYQCSGKGSN 321
Query: 476 VANRVKWRGYHVLNNSQAMDFTVNQFIKGDLWL 508
RV+W L++ +A F ++I GD WL
Sbjct: 322 RTGRVEWS--KSLSSEEARPFLGREYISGDEWL 414
>BP039949
Length = 517
Score = 90.9 bits (224), Expect = 5e-19
Identities = 52/145 (35%), Positives = 79/145 (53%), Gaps = 2/145 (1%)
Frame = -3
Query: 366 VFQNCLIQP--KKPLKDQKNTIAAQSRTHPNQTSGFSFHNCTIIPDPDFQPSNGSIQTFL 423
+++ C I K L +I AQ R N+ +GFSF NCTI+ G+ +L
Sbjct: 515 LYEGCTIHSIAKPDLDGVSGSITAQGRQSINEETGFSFVNCTIL---------GTGNVWL 363
Query: 424 GRPWKNFSRTVFMESFMSEMIHPEGWLEWETNTTYDDTLFFGEYQNKGPGSGVANRVKWR 483
GR W FS VF +++MSE++ PEGW +W + + D T+FFGEY+ GPG+ +RV +
Sbjct: 362 GRAWGAFSTAVFSKTYMSEVVAPEGWNDWR-DPSRDQTVFFGEYRCLGPGANYTSRVSYA 186
Query: 484 GYHVLNNSQAMDFTVNQFIKGDLWL 508
L + +A + +I G+ WL
Sbjct: 185 --KQLKDYEANSYMNISYINGNDWL 117
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,628,046
Number of Sequences: 28460
Number of extensions: 140023
Number of successful extensions: 773
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 735
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 735
length of query: 521
length of database: 4,897,600
effective HSP length: 94
effective length of query: 427
effective length of database: 2,222,360
effective search space: 948947720
effective search space used: 948947720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0343.3


