

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
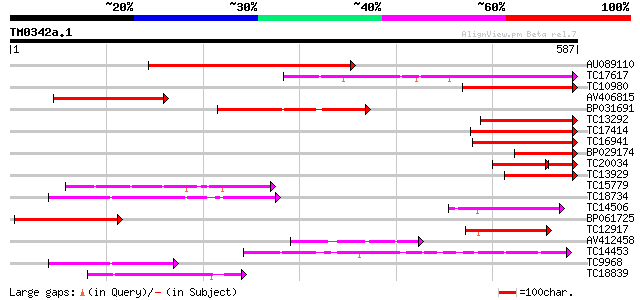
Query= TM0342a.1
(587 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AU089110 426 e-120
TC17617 weakly similar to UP|Q84SA8 (Q84SA8) Laccase (Fragment),... 255 1e-68
TC10980 similar to UP|Q8VZA1 (Q8VZA1) Laccase (Diphenol oxidase)... 199 8e-52
AV406815 157 3e-39
BP031691 148 3e-36
TC13292 similar to UP|Q9FI28 (Q9FI28) Laccase (Diphenol oxidase)... 146 8e-36
TC17414 weakly similar to UP|Q84SA8 (Q84SA8) Laccase (Fragment),... 142 2e-34
TC16941 similar to UP|Q8L4Y3 (Q8L4Y3) Diphenol oxidase laccase, ... 137 4e-33
BP029174 122 1e-28
TC20034 weakly similar to UP|Q9LM34 (Q9LM34) T10O22.11, partial ... 91 3e-26
TC13929 weakly similar to UP|Q9AUI0 (Q9AUI0) Laccase, partial (14%) 114 6e-26
TC15779 similar to UP|O24093 (O24093) L-ascorbate oxidase precur... 112 2e-25
TC18734 similar to UP|Q8LFM3 (Q8LFM3) Pectinesterase-like protei... 107 4e-24
TC14506 homologue to UP|O24093 (O24093) L-ascorbate oxidase prec... 101 4e-22
BP061725 93 1e-19
TC12917 similar to UP|Q9M5B5 (Q9M5B5) Ascorbate oxidase AO1, par... 91 4e-19
AV412458 89 2e-18
TC14453 similar to PIR|T07634|T07634 pollen-specific protein hom... 85 3e-17
TC9968 similar to UP|Q40473 (Q40473) PS60 protein precursor, par... 84 5e-17
TC18839 similar to GB|BAB01744.1|9280289|AP000603 l-ascorbate ox... 79 2e-15
>AU089110
Length = 646
Score = 426 bits (1094), Expect = e-120
Identities = 210/215 (97%), Positives = 210/215 (97%)
Frame = +1
Query: 144 TLYGSLIILPKHNAQYPFVKPYKEVPMIFGEWFNADPEAIISQALQTGGGPNVSDAYTIN 203
T YG LIILPKHNAQYPFVKPYKEVPMIFG WFNADPEAIISQALQTGGGPNVSDAYTIN
Sbjct: 1 TSYGPLIILPKHNAQYPFVKPYKEVPMIFGXWFNADPEAIISQALQTGGGPNVSDAYTIN 180
Query: 204 GLPGPLYNCSKKDTFKLKVKPGKIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPF 263
GLPGPLYNCSKKDTFKLKVKPG IYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPF
Sbjct: 181 GLPGPLYNCSKKDTFKLKVKPGXIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPF 360
Query: 264 VTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQHNIT 323
VTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGL TFDNSTVAGILEYKIQHNIT
Sbjct: 361 VTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLXTFDNSTVAGILEYKIQHNIT 540
Query: 324 HHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLR 358
HHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLR
Sbjct: 541 HHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLR 645
>TC17617 weakly similar to UP|Q84SA8 (Q84SA8) Laccase (Fragment), partial
(53%)
Length = 1139
Score = 255 bits (652), Expect = 1e-68
Identities = 141/316 (44%), Positives = 190/316 (59%), Gaps = 12/316 (3%)
Frame = +2
Query: 284 SHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQHNITHHNSAVKLKKLPLFKPLLPA 343
++ P +++M A PY +G FDN T GI+ Y Q+ + +S + + LP P++P
Sbjct: 20 ANQPIGSYYMVASPYKSGGKMFDNITSRGIVVYDHQY-AQYSSSHLVMPSLPSLNPMMPF 196
Query: 344 L---NDTSFATKFSNKLRSLASARFPANVPQKVDKHFLFTVGLGTNPCK--NKSNQTCQG 398
L NDT A KF + + SL A VP +VD+H TVGL C +N TC+G
Sbjct: 197 LPPFNDTPTAHKFYSNITSLVGAPHWVPVPLEVDEHMFITVGLNLQRCDPATVTNDTCKG 376
Query: 399 PNNATMFSGSVNNVSFILPTT---ALLQAHFFGKSNGVYTPDFPTSPLHPFNYTGTTP-- 453
P N T+ S S+NN SF+LP ++L+A FF +GVYT DFP+ P F++T
Sbjct: 377 PMNHTL-SPSMNNESFVLPKGRGYSMLEA-FFENMSGVYTTDFPSKPPVMFDFTNPDIRF 550
Query: 454 --NNTNVSNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNK 511
N TKV L FN++VE+V Q+T+ + ++H +HLHGFNF V+ QGFGNFDP
Sbjct: 551 DLNLIFAPKSTKVKKLKFNSTVEIVFQNTAFIHTDNHAMHLHGFNFHVLAQGFGNFDPTS 730
Query: 512 DPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGK 571
D + FNLV+P RNT+ VP GGW IRF A+NPGVWF+HCH+E HT G MA+ V +G
Sbjct: 731 DEAKFNLVNPQIRNTIAVPVGGWAVIRFQANNPGVWFVHCHVEHHTHRGFNMAFEVENGP 910
Query: 572 LPNQKLLPPPVDLPKC 587
+ L PPP DLPKC
Sbjct: 911 TLSTSLTPPPADLPKC 958
>TC10980 similar to UP|Q8VZA1 (Q8VZA1) Laccase (Diphenol oxidase)-like
protein, partial (22%)
Length = 572
Score = 199 bits (507), Expect = 8e-52
Identities = 87/119 (73%), Positives = 101/119 (84%)
Frame = +1
Query: 469 FNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVG 528
FN++VELV+QDT++L ESHP HLHG+NFFVVG G GNFDP+KDP+ +NLVDPVERNTVG
Sbjct: 7 FNSTVELVLQDTNLLSVESHPFHLHGYNFFVVGTGVGNFDPSKDPAKYNLVDPVERNTVG 186
Query: 529 VPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
VP+GGW AIRF ADNPGVWF HCHLE+HT WGLK A+VV DG +Q +LPPP DLP C
Sbjct: 187 VPTGGWTAIRFRADNPGVWFFHCHLELHTGWGLKTAFVVEDGPGQDQSVLPPPKDLPAC 363
>AV406815
Length = 426
Score = 157 bits (398), Expect = 3e-39
Identities = 69/119 (57%), Positives = 88/119 (72%)
Frame = +2
Query: 46 VTRLCHSKSMVTVNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSGWADGP 105
V RLC ++ ++TVNG+FPGP + R GD + IKV N NISIHWHG+R L++ WADGP
Sbjct: 68 VKRLCRTQRILTVNGQFPGPTVQVRNGDSVAIKVTNAGPYNISIHWHGLRMLRNPWADGP 247
Query: 106 AYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGSLIILPKHNAQYPFVKP 164
+YVTQCPIQ G SY Y +TI+ Q GTL+WHAH +LRAT+YG+ II P+ + YPF P
Sbjct: 248 SYVTQCPIQPGGSYTYRFTIQNQEGTLWWHAHTGFLRATVYGAFIIYPRLGSPYPFSMP 424
>BP031691
Length = 481
Score = 148 bits (373), Expect = 3e-36
Identities = 82/162 (50%), Positives = 107/162 (65%), Gaps = 4/162 (2%)
Frame = +1
Query: 216 DTFKLKVKPGKIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPFVTKTILITPGQT 275
DTF V+ GK YLLR+INAAL+DEL+F +ANHTL VVE DA Y KPF T+ I++TPGQT
Sbjct: 13 DTFIYTVESGKTYLLRIINAALSDELYFGVANHTLTVVEVDASYTKPFNTRAIMVTPGQT 192
Query: 276 TNVLLKTKSHYPNAT--FFMTARPYVTGLGTFDNSTVAGILEYKIQHNITHHNSAVKLKK 333
TNVLL+ + P+++ F M ARPY T + FDNST G L Y + VK
Sbjct: 193 TNVLLRA-NQIPDSSGLFVMAARPYRTSVFPFDNSTTIGYLRYN-----STRGEKVKPPH 354
Query: 334 LPLFKPL--LPALNDTSFATKFSNKLRSLASARFPANVPQKV 373
+P + LP + DT F T+FS KLRS+A+++FP VP+K+
Sbjct: 355 VPTDLSIHNLPEMEDTKFETEFSAKLRSIATSQFPCKVPKKI 480
>TC13292 similar to UP|Q9FI28 (Q9FI28) Laccase (Diphenol oxidase), partial
(19%)
Length = 558
Score = 146 bits (369), Expect = 8e-36
Identities = 62/100 (62%), Positives = 76/100 (76%)
Frame = +2
Query: 488 HPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVW 547
HP+HLHGF+F VVG GFGNF+ K P ++L+DP NTV VP GW AIRF+ADNPGVW
Sbjct: 8 HPMHLHGFSFHVVGYGFGNFNKRKSPMYYDLIDPPLLNTVIVPKNGWAAIRFVADNPGVW 187
Query: 548 FMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
FMHCHLE H SWG++ ++V +GK + KLLPPP D+P C
Sbjct: 188 FMHCHLERHLSWGMETVFIVKNGKSSHAKLLPPPPDMPPC 307
>TC17414 weakly similar to UP|Q84SA8 (Q84SA8) Laccase (Fragment), partial
(27%)
Length = 670
Score = 142 bits (357), Expect = 2e-34
Identities = 59/110 (53%), Positives = 80/110 (72%)
Frame = +3
Query: 478 QDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAI 537
Q+T++L A+SHP+H+HG+NF V+ Q FG ++P D + +NLV+P TV VP GGW AI
Sbjct: 15 QNTAVLNAQSHPMHIHGYNFHVLAQDFGIYNPILDKAKYNLVNPQISTTVPVPPGGWAAI 194
Query: 538 RFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
RF A+NPG WF+HCH++ H WGL ++V +G P+ LLPPP DLPKC
Sbjct: 195 RFQANNPGAWFVHCHVDDHNVWGLTTVFLVENGPTPSTSLLPPPADLPKC 344
>TC16941 similar to UP|Q8L4Y3 (Q8L4Y3) Diphenol oxidase laccase, partial
(18%)
Length = 625
Score = 137 bits (346), Expect = 4e-33
Identities = 60/108 (55%), Positives = 76/108 (69%)
Frame = +2
Query: 480 TSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRF 539
TSI+ E+HP+HLHG++F+VV +GFGNF+P K S FNL+DP R TV VP+ GW IRF
Sbjct: 2 TSIVTPENHPIHLHGYDFYVVAEGFGNFNPKKHTSKFNLLDPPMRYTVSVPANGWAVIRF 181
Query: 540 LADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
+ADNPG W MHCHL+VH WGL +V +G + + PP DLP C
Sbjct: 182 VADNPGAWIMHCHLDVHIGWGLATILLVDNGVGLLESIESPPEDLPLC 325
>BP029174
Length = 493
Score = 122 bits (307), Expect = 1e-28
Identities = 52/65 (80%), Positives = 59/65 (90%)
Frame = -2
Query: 523 ERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKLPNQKLLPPPV 582
ERNTVGVP+GGW AIRF ADNPGVWFMHCHLE+HT+WGLKMA+VV +GK PN+ +LPPP
Sbjct: 492 ERNTVGVPAGGWTAIRFRADNPGVWFMHCHLEIHTTWGLKMAFVVDNGKGPNESILPPPR 313
Query: 583 DLPKC 587
DLPKC
Sbjct: 312 DLPKC 298
>TC20034 weakly similar to UP|Q9LM34 (Q9LM34) T10O22.11, partial (15%)
Length = 457
Score = 90.9 bits (224), Expect(2) = 3e-26
Identities = 38/60 (63%), Positives = 43/60 (71%)
Frame = +2
Query: 501 GQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWG 560
G GFGNFD KDP +NLVDP +NTV +P GW IRF A NPGVWFMHCH+E H + G
Sbjct: 2 GWGFGNFD*EKDPLGYNLVDPPYQNTVAIPKNGWTTIRFRAKNPGVWFMHCHIERHVTLG 181
Score = 45.1 bits (105), Expect(2) = 3e-26
Identities = 15/29 (51%), Positives = 23/29 (78%)
Frame = +3
Query: 559 WGLKMAWVVLDGKLPNQKLLPPPVDLPKC 587
WG+ M ++V DG P +K+LPPP+D+P+C
Sbjct: 177 WGMAMTFIVKDGNNPEEKMLPPPLDMPQC 263
>TC13929 weakly similar to UP|Q9AUI0 (Q9AUI0) Laccase, partial (14%)
Length = 484
Score = 114 bits (284), Expect = 6e-26
Identities = 45/75 (60%), Positives = 60/75 (80%)
Frame = +1
Query: 513 PSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGKL 572
P+ FNLVDP +NT+G P GGWVAIRF+ADNPG+WF+HCH++ H +WGL A +V +G
Sbjct: 1 PARFNLVDPPVKNTIGTPPGGWVAIRFVADNPGIWFLHCHIDSHLNWGLATALLVENGVG 180
Query: 573 PNQKLLPPPVDLPKC 587
P+Q ++PPP DLP+C
Sbjct: 181 PSQSVIPPPPDLPQC 225
>TC15779 similar to UP|O24093 (O24093) L-ascorbate oxidase precursor,
partial (50%)
Length = 1304
Score = 112 bits (279), Expect = 2e-25
Identities = 87/245 (35%), Positives = 122/245 (49%), Gaps = 27/245 (11%)
Frame = +1
Query: 58 VNGEFPGPHIVAREGDRLIIKVVN--HVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQT 115
+NG+FPGP I A GD L I + N H + + IH HGIRQL + WADG A ++QC I
Sbjct: 265 INGQFPGPTIKAEVGDTLDIALTNKLHTEGTV-IHGHGIRQLGTPWADGTAAISQCAINP 441
Query: 116 GQSYVYNYTIKGQRGTLFWHAHISWLRAT-LYGSLII-LPKHNAQYPFVKPYKEVPMIFG 173
G+++ Y +T+ + GT F+H H RA LYGSLI+ LPK + PF + ++
Sbjct: 442 GETFHYWFTV-DRPGTYFYHGHYGMQRAAGLYGSLIVDLPKGQNE-PFHYDGEFNLLLSD 615
Query: 174 EWFNADPE---AIISQALQTGGGPNVSDAYTINGLPGPLYNCSKKDTFK----------- 219
W + E + S+ L+ G P + ING +NCS F
Sbjct: 616 LWHKSSHEQEVGLSSKPLKWIGEP---QSLLING--RGQFNCSLAAKFVNTSLVPQCQFK 780
Query: 220 ---------LKVKPGKIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPFVTKTILI 270
L V+P K Y +R+ + L +I+NH L VVEAD +V+PF I I
Sbjct: 781 GGEECAPEILHVEPNKTYRIRIASTTSMASLNLAISNHKLIVVEADGNHVQPFAVDDIDI 960
Query: 271 TPGQT 275
G+T
Sbjct: 961 YSGET 975
>TC18734 similar to UP|Q8LFM3 (Q8LFM3) Pectinesterase-like protein, partial
(47%)
Length = 929
Score = 107 bits (268), Expect = 4e-24
Identities = 65/241 (26%), Positives = 115/241 (46%), Gaps = 1/241 (0%)
Frame = +3
Query: 41 IRYQNVTRLCHSKSMVTVNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSG 100
+ Y ++ L + ++ +NG+FPGP + D +I+ ++N + + W+GI+Q ++
Sbjct: 102 VTYGTLSPLGSPQQVILINGQFPGPRLDLVTNDNVILNLINKLDEPFLLTWNGIKQRKNS 281
Query: 101 WADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT-LYGSLIILPKHNAQY 159
W DG T CPI +Y Y + K Q GT + +A+ +G+L +L +
Sbjct: 282 WQDG-VLGTNCPIPPNSNYTYKFQAKDQIGTYTYFPSTKMHKASGGFGALNVLHRSVIPI 458
Query: 160 PFVKPYKEVPMIFGEWFNADPEAIISQALQTGGGPNVSDAYTINGLPGPLYNCSKKDTFK 219
P+ P + ++ G+W+ + +SQ L +G D ING TF
Sbjct: 459 PYPNPDGDFTLLIGDWYKTSHKT-LSQTLDSGKSLAFPDGLLING--------QAHSTF- 608
Query: 220 LKVKPGKIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPFVTKTILITPGQTTNVL 279
V GK Y+ R+ N L+ + F I HTLK+VE + + ++ + GQ+ +VL
Sbjct: 609 -TVNQGKTYMFRISNVGLSTSINFRIQGHTLKLVEVEGSHTIQNEYDSLDVHVGQSVSVL 785
Query: 280 L 280
+
Sbjct: 786 V 788
>TC14506 homologue to UP|O24093 (O24093) L-ascorbate oxidase precursor,
partial (24%)
Length = 692
Score = 101 bits (251), Expect = 4e-22
Identities = 52/123 (42%), Positives = 71/123 (57%), Gaps = 3/123 (2%)
Frame = +3
Query: 455 NTNVSNGTKVVVLPFNTSVELVMQDTSIL---GAESHPLHLHGFNFFVVGQGFGNFDPNK 511
NT V NG V + N V++++Q+ + L G+E HP HLHG +F+V+G G G F P
Sbjct: 30 NTTVGNG--VYMFQLNEVVDVILQNANQLNGNGSEIHPWHLHGHDFWVLGYGEGKFRPGV 203
Query: 512 DPSNFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLKMAWVVLDGK 571
D +FNL RNT + GW A+RF ADNPGVW HCH+E H G+ + + K
Sbjct: 204 DEKSFNLTRAPLRNTAVIFPYGWTALRFKADNPGVWAFHCHIEPHLHMGMGVIFAEAVHK 383
Query: 572 LPN 574
+ N
Sbjct: 384 VKN 392
>BP061725
Length = 358
Score = 92.8 bits (229), Expect = 1e-19
Identities = 42/111 (37%), Positives = 68/111 (60%)
Frame = +3
Query: 6 RLKIYTLLYLFISLCYILFYSFVCSSCNKIRLLTQIRYQNVTRLCHSKSMVTVNGEFPGP 65
R+ ++ LF+ + LF + + + ++R + T+LC +K+++TVNG+FPGP
Sbjct: 21 RIYSSSMKILFLQVLLFLFINLLLTCLALRHHKFEVREVSYTKLCSTKNILTVNGQFPGP 200
Query: 66 HIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTG 116
+ +G+ +I+ V N NI+IH HG+ Q + W+DGP YVTQCPIQ G
Sbjct: 201 TLYVTKGETIIVHVYNRANYNITIHXHGVNQPRYPWSDGPEYVTQCPIQPG 353
>TC12917 similar to UP|Q9M5B5 (Q9M5B5) Ascorbate oxidase AO1, partial (19%)
Length = 521
Score = 91.3 bits (225), Expect = 4e-19
Identities = 42/93 (45%), Positives = 60/93 (64%), Gaps = 4/93 (4%)
Frame = +1
Query: 473 VELVMQDTSILG----AESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVG 528
V++V+Q+T+IL ++ HP HLHG +F+++G G G F P D + FNL +P RN
Sbjct: 1 VDVVLQNTNILERIKRSDFHPWHLHGHDFWILGYGDGMFKPGDDETKFNLENPPLRNNAV 180
Query: 529 VPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGL 561
+ GW A+RF A+NPGVW HCH+E H G+
Sbjct: 181 LFPYGWTALRFKANNPGVWAFHCHVEPHLHMGM 279
>AV412458
Length = 423
Score = 89.0 bits (219), Expect = 2e-18
Identities = 55/140 (39%), Positives = 80/140 (56%), Gaps = 2/140 (1%)
Frame = +2
Query: 291 FFMTARPYVTGLGT-FDNSTVAGILEYKIQHNITHHNSAVKLKKLPLFKPLLPALNDTSF 349
++M AR Y T + FDN+T ILEY ++ ++ + PLLPA NDT
Sbjct: 23 YYMAARAYQTAMNAAFDNTTTTAILEYIPRNRNVQPSTPIL--------PLLPAFNDTPT 178
Query: 350 ATKFSNKLRSLASARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNATMFSGSV 409
AT F++ +R L+ N D + FTVGLG C+N ++ CQGP N + F+ S+
Sbjct: 179 ATAFTSGIRGLSQNVVFKN---SADVNLFFTVGLGLIFCRNPNSPNCQGP-NGSRFAASI 346
Query: 410 NNVSFILP-TTALLQAHFFG 428
NN SF+LP TT+L+QA++ G
Sbjct: 347 NNNSFVLPRTTSLMQAYYQG 406
>TC14453 similar to PIR|T07634|T07634 pollen-specific protein homolog
T1P17.10 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (56%)
Length = 1364
Score = 85.1 bits (209), Expect = 3e-17
Identities = 95/344 (27%), Positives = 140/344 (40%), Gaps = 5/344 (1%)
Frame = +2
Query: 243 FSIANHTLKVVEADAVYVKPFVTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGL 302
F I NH L + E + Y ++ + GQ+ L+ T + +++ A
Sbjct: 2 FRIQNHNLLLAETEGSYTVQQNYTSLDVHVGQSYTFLV-TMDQNASTDYYIVASARFVNE 178
Query: 303 GTFDNSTVAGILEYKIQHNITHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSL-- 360
+ T GIL Y NS K + PL A D T N+ RS+
Sbjct: 179 SRWQRVTGVGILHYT--------NSKGKARG-----PLPAAPEDQFDKTYSMNQARSIRW 319
Query: 361 -ASARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNATMFSGSVNNVSFILPTT 419
SA PQ ++ V KNK G AT+ + +SF+ P+T
Sbjct: 320 NVSASGARPNPQGSFRYGSINV-TEVYVLKNKPRVKINGKKRATL-----SGISFVNPST 481
Query: 420 ALLQAHFFGKSNGVYTPDFPTSPLHPFNYTGTTPNNTNVSNGTKVVVLPFNTSVELVMQD 479
+ A F K +Y DFPT PL TG+ T+V NG+ + +E+++Q+
Sbjct: 482 PIRLADQF-KLKDIYKLDFPTRPL-----TGSPRAETSVINGS------YRGFIEIILQN 625
Query: 480 TSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSNFNLVDPVERNTVGVPSGGWVAIRF 539
H HL G+ FFVVG +G++ N +N D + R T V G W AI
Sbjct: 626 ND---TRMHTYHLDGYAFFVVGMDYGDWSDNSR-GTYNKWDGIARATTQVYPGAWTAILV 793
Query: 540 LADNPGVWFMHCHLEVHTSWGL-KMAWVVLDGKLPNQKL-LPPP 581
DN G+W + E SW L + ++ + PN K LP P
Sbjct: 794 SLDNVGIW--NLRTENLDSWYLGQETYIRVVNPEPNNKTELPIP 919
>TC9968 similar to UP|Q40473 (Q40473) PS60 protein precursor, partial (27%)
Length = 631
Score = 84.3 bits (207), Expect = 5e-17
Identities = 41/135 (30%), Positives = 76/135 (55%), Gaps = 1/135 (0%)
Frame = +2
Query: 41 IRYQNVTRLCHSKSMVTVNGEFPGPHIVAREGDRLIIKVVNHVQNNISIHWHGIRQLQSG 100
I Y ++ L + + +NG+FPGP I + D L+I V NH+ + W+G++Q ++
Sbjct: 230 ITYGDIYPLGVKQQGILINGQFPGPEIYSVTNDNLVINVHNHLPEPFLLSWNGVQQRRNS 409
Query: 101 WADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT-LYGSLIILPKHNAQY 159
+ DG Y T CPI G+++ YN +K Q G+ F+ +++ +A +G++ IL +
Sbjct: 410 YQDG-VYGTTCPIPPGKNFTYNLQVKDQIGSFFYFPSLAFHKAAGGFGAIKILSRPRIPV 586
Query: 160 PFVKPYKEVPMIFGE 174
PF +P + ++ G+
Sbjct: 587 PFPEPAADFSLLIGD 631
>TC18839 similar to GB|BAB01744.1|9280289|AP000603 l-ascorbate oxidase;
pectinesterase-like protein; pollen-specific
protein-like {Arabidopsis thaliana;} , partial (29%)
Length = 498
Score = 79.0 bits (193), Expect = 2e-15
Identities = 53/173 (30%), Positives = 78/173 (44%), Gaps = 8/173 (4%)
Frame = +1
Query: 81 NHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISW 140
N++ + WHG++Q ++ W DG A T CPI G +Y Y++ +K Q G+ F++ +
Sbjct: 13 NNLDEPLQFTWHGVQQRKNSWQDGVAG-TSCPILPGTNYTYHFQVKDQIGSYFYYPTTAL 189
Query: 141 LRAT-LYGSLIILPKHNAQYPFVKPYKEVPMIFGEWFNADPEAIISQALQTGGGPNVSDA 199
RA +G L I + P+ P E +I G+W+ A + L +G D
Sbjct: 190 HRAAGGFGGLRIFSRLLIPVPYPDPEDEYWVIIGDWY-AKSHKTLKSFLDSGRSIGRPDG 366
Query: 200 YTINGLPG-------PLYNCSKKDTFKLKVKPGKIYLLRLINAALNDELFFSI 245
ING PLY +KPGK Y R+ N L D L F I
Sbjct: 367 VLINGKTAKGDGSDQPLYT----------MKPGKTYKYRICNVGLKDSLNFRI 495
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,958,671
Number of Sequences: 28460
Number of extensions: 187239
Number of successful extensions: 1222
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 1199
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1205
length of query: 587
length of database: 4,897,600
effective HSP length: 95
effective length of query: 492
effective length of database: 2,193,900
effective search space: 1079398800
effective search space used: 1079398800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0342a.1


