

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0333b.9
(488 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
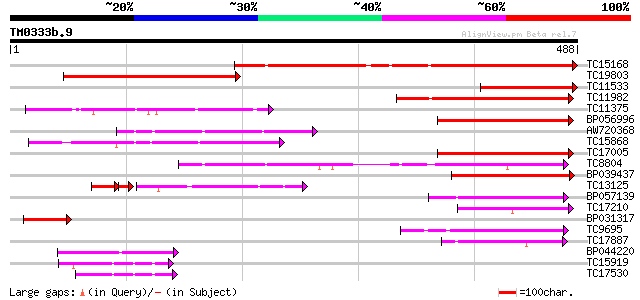
Score E
Sequences producing significant alignments: (bits) Value
TC15168 similar to UP|O04084 (O04084) Serine carboxypeptidase is... 286 7e-78
TC19803 similar to UP|O65568 (O65568) Serine carboxypeptidase II... 196 5e-51
TC11533 similar to UP|Q9LSM9 (Q9LSM9) Serine carboxypeptidase II... 174 4e-44
TC11982 similar to UP|Q9M099 (Q9M099) Serine carboxypeptidase II... 155 1e-38
TC11375 similar to UP|Q8L7B2 (Q8L7B2) Serine carboxypeptidase 1-... 135 1e-32
BP056996 135 1e-32
AW720368 132 2e-31
TC15868 weakly similar to UP|KEX1_YEAST (P09620) Carboxypeptidas... 130 6e-31
TC17005 similar to UP|CP22_HORVU (P55748) Serine carboxypeptidas... 128 2e-30
TC8804 similar to UP|Q8L6A7 (Q8L6A7) Carboxypeptidase type III, ... 115 2e-26
BP039437 108 3e-24
TC13125 similar to PIR|C96759|C96759 protein serine carboxypepti... 75 2e-19
BP057139 87 8e-18
TC17210 similar to UP|Q9FH05 (Q9FH05) Serine carboxypeptidase II... 84 5e-17
BP031317 83 9e-17
TC9695 similar to UP|Q8L7B2 (Q8L7B2) Serine carboxypeptidase 1-l... 82 3e-16
TC17887 weakly similar to UP|AAQ91191 (AAQ91191) 1-O-sinapoylglu... 75 2e-14
BP044220 69 2e-12
TC15919 similar to UP|Q9XH61 (Q9XH61) Serine carboxypeptidase, p... 67 7e-12
TC17530 similar to UP|Q8L6A7 (Q8L6A7) Carboxypeptidase type III,... 65 3e-11
>TC15168 similar to UP|O04084 (O04084) Serine carboxypeptidase isolog,
30227-33069, partial (55%)
Length = 1005
Score = 286 bits (731), Expect = 7e-78
Identities = 141/297 (47%), Positives = 192/297 (64%), Gaps = 2/297 (0%)
Frame = +1
Query: 194 SGESYGGHYIPQLAELIFDRNKDKNKYPFINLKGFIVGNPETEDYYDYKGLLEYAWSHAV 253
+GESY G Y+P+LAELI DRNKD + + I+LKG ++GNPET D D+ GL++YAWSHAV
Sbjct: 1 AGESYAGKYVPELAELILDRNKDMSLH--IDLKGILLGNPETSDAEDWMGLVDYAWSHAV 174
Query: 254 ISDQQYDKAKQVCDFKQFQ-WSNE-CNKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIAS 311
ISD+ + K+ CDF W NE C++A++EV + Y EIDIY++Y C ++ + S
Sbjct: 175 ISDETHQTVKKSCDFNSSDPWHNEDCSQAVDEVLKQYKEIDIYSLYTSTCFASTAN---S 345
Query: 312 EGLEQLTKGRNDYRMKRKRIFGGYDPCYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEV 371
G T + M R+ GGYDPC YA+ ++NR DVQ + H + G+ W +
Sbjct: 346 NGQSVQTSMKRSSTMM-PRMMGGYDPCLDDYAKTFYNRPDVQKALHASD--GHNLKNWSI 516
Query: 372 CNNSILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSS 431
CN++I + S S++PIY KLI GLKIW+YSGD DGRVPV+ TRY + +L LP+
Sbjct: 517 CNHNIFNNWGDSKPSVIPIYKKLISAGLKIWVYSGDTDGRVPVLSTRYSLSSLALPVTKP 696
Query: 432 WRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLPTRR 488
WR WY DN+V G EY+GLT+ T RGAGH VP KP +L+ SFL G+ P+ +
Sbjct: 697 WRPWYHDNEVSGWFEEYQGLTFATFRGAGHAVPCFKPSNSLAFFSSFLLGESPPSTK 867
>TC19803 similar to UP|O65568 (O65568) Serine carboxypeptidase II - like
protein (Serine carboxypeptidase II-like protein),
partial (36%)
Length = 593
Score = 196 bits (499), Expect = 5e-51
Identities = 90/152 (59%), Positives = 112/152 (73%)
Frame = +2
Query: 47 DRIVDLPGVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGCSS 106
D++ LPG + S +H+SGYITVN+ GRALFYWF EA +P KPLLLWLNGGPGCSS
Sbjct: 134 DKVGRLPGQNFNTSFAHYSGYITVNDKAGRALFYWFMEADQDPQSKPLLLWLNGGPGCSS 313
Query: 107 VGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILEDN 166
+ YG A E+GP + +G+ L NPY+WNQ AN+L V++P GVGFSY+NTSSDL D
Sbjct: 314 IAYGEAEEMGPFHIKPDGKTLYLNPYAWNQVANILSVDAPAGVGFSYSNTSSDLLNHGDK 493
Query: 167 FVAEDAYNFLVNWLQRFPQFKSRGFFISGESY 198
AED+ FL+ W +RFPQ+K R FFI+GESY
Sbjct: 494 KTAEDSLIFLLKWFERFPQYKGRDFFITGESY 589
>TC11533 similar to UP|Q9LSM9 (Q9LSM9) Serine carboxypeptidase II-like
protein, partial (17%)
Length = 526
Score = 174 bits (440), Expect = 4e-44
Identities = 83/83 (100%), Positives = 83/83 (100%)
Frame = +3
Query: 406 GDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPL 465
GDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPL
Sbjct: 3 GDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPL 182
Query: 466 NKPKEALSLIHSFLAGDRLPTRR 488
NKPKEALSLIHSFLAGDRLPTRR
Sbjct: 183 NKPKEALSLIHSFLAGDRLPTRR 251
>TC11982 similar to UP|Q9M099 (Q9M099) Serine carboxypeptidase II-like
protein, partial (33%)
Length = 701
Score = 155 bits (393), Expect = 1e-38
Identities = 77/152 (50%), Positives = 94/152 (61%)
Frame = +2
Query: 334 GYDPCYSTYAEKYFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTK 393
GYDPC YAEKY+NR DVQ + H N N W C++ + + + S SILPIY +
Sbjct: 41 GYDPCTENYAEKYYNRYDVQKAMHANVT--NIPYKWTACSDVLNKHWKDSEVSILPIYKE 214
Query: 394 LIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYEGLTY 453
LI GL+IW++SGD D VPV TR+ + L L +K+ W WY QVGG Y GLT+
Sbjct: 215 LIAAGLRIWVFSGDTDSVVPVTATRFSLNHLNLAIKTRWYPWYSGVQVGGWTEVYNGLTF 394
Query: 454 VTVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
TVRGAGH VPL +PK A L SFLAG LP
Sbjct: 395 ATVRGAGHEVPLFQPKRAYILFRSFLAGKELP 490
>TC11375 similar to UP|Q8L7B2 (Q8L7B2) Serine carboxypeptidase 1-like
protein (Serine carboxypeptidase 1 precursor-like
protein), partial (35%)
Length = 874
Score = 135 bits (341), Expect = 1e-32
Identities = 82/223 (36%), Positives = 124/223 (54%), Gaps = 9/223 (4%)
Frame = +3
Query: 14 NMNVILSVHFLCFFLLTTSFIKTY-GINVGANESDRIVDLPGVPSSPSVSHFSGYITV-- 70
N + +L+ LC +L I V A S + G + PS H+SGY+ +
Sbjct: 84 NYHKLLATSSLCIMILFDHLIGVVEAAPVQALSSQNSLCFKG--NFPS-KHYSGYVNIDG 254
Query: 71 NEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGCSSVGYGAAIEIGPL---LVNKNGE-- 125
N ++G+ LFY+F ++ P K P++LWLNGGPGCSS+ G E GP G+
Sbjct: 255 NGENGKNLFYYFVSSERNPGKDPVVLWLNGGPGCSSLD-GFVYEHGPFNFQAAKTKGDLP 431
Query: 126 GLNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQ 185
L+ NPYSW++ +N+++++SP GVG SY+ SS + +D A D + FL W ++FP+
Sbjct: 432 ILHINPYSWSKVSNIIYLDSPAGVGLSYSKNSSKY-VTDDLQTASDTHAFLFKWFEQFPE 608
Query: 186 FKSRGFFISGESYGGHYIPQLA-ELIFDRNKDKNKYPFINLKG 227
F + F++SGESY G Y+P L+ E++ K P IN G
Sbjct: 609 FLANPFYLSGESYAGVYVPTLSIEVV--XGIXSGKKPLINFNG 731
>BP056996
Length = 550
Score = 135 bits (341), Expect = 1e-32
Identities = 60/117 (51%), Positives = 79/117 (67%)
Frame = -2
Query: 369 WEVCNNSILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPL 428
W C++++ +N + S LP+ KLI GGL++W+YSGD DGR+PV TRY ++ LGL +
Sbjct: 525 WSHCSDNVSNFWNVAPQSTLPVIKKLIAGGLRVWVYSGDTDGRIPVTSTRYTLKKLGLKI 346
Query: 429 KSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
W WY QVGG VEY+GLT+VT+RGAGH VP PK+AL LI FL +LP
Sbjct: 345 VEDWTPWYTSRQVGGWTVEYDGLTFVTIRGAGHQVPTFAPKQALQLIRHFLVDKKLP 175
>AW720368
Length = 570
Score = 132 bits (331), Expect = 2e-31
Identities = 73/173 (42%), Positives = 98/173 (56%)
Frame = +2
Query: 93 PLLLWLNGGPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVESPVGVGFS 152
P+++WL GGPGCSS E GP KN L +N Y W++ +N+LFV+ P G GFS
Sbjct: 20 PVVIWLTGGPGCSSE-LAMFYENGPFHFAKN-LSLVWNEYGWDKASNILFVDQPTGTGFS 193
Query: 153 YTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFD 212
YT SD+ E+ V+ D Y+FL + + QF FFI+GESY GHYIP LA +
Sbjct: 194 YTTDESDIRHDEEG-VSNDLYDFLQAFFKEHSQFTKNDFFITGESYAGHYIPALASRVHQ 370
Query: 213 RNKDKNKYPFINLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQV 265
NK K + INLKGF +GN T YK +YA +I+ YD+ ++
Sbjct: 371 GNKAK-EGTHINLKGFAIGNGLTNPEIQYKAYTDYALDMGLINKTDYDRINKL 526
>TC15868 weakly similar to UP|KEX1_YEAST (P09620) Carboxypeptidase KEX1
precursor (Carboxypeptidase D) , partial (8%)
Length = 780
Score = 130 bits (326), Expect = 6e-31
Identities = 77/223 (34%), Positives = 120/223 (53%), Gaps = 3/223 (1%)
Frame = +2
Query: 17 VILSVHFLCFFLLTTSFIKTYGINVGANESDRIVDLPGVPSSPSVSHFSGYITVNEDHGR 76
+I ++ F CFF S ++ + ++ P + ++ SGY+ V+
Sbjct: 44 IIKNLIFFCFFFFQISHSHSHSTSTPRSK-------PHIFPKEALPTKSGYLPVDPTTTS 202
Query: 77 ALFYWFFEAQSEPS---KKPLLLWLNGGPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYS 133
++FY F+EAQ+ S + PLL+WL GGPGCSS+ G E+GP V N L NP S
Sbjct: 203 SIFYTFYEAQNSTSPLSQTPLLIWLQGGPGCSSM-IGNFYELGPWRVT-NSLTLEPNPGS 376
Query: 134 WNQEANLLFVESPVGVGFSYTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFI 193
WN+ LLF++SP+G GFS T ++ + VA+ Y + ++Q P FK R +I
Sbjct: 377 WNRIYGLLFLDSPIGTGFSVGTTPEEIPA-DQTGVAKHLYAAIKGFIQLDPVFKDRPIYI 553
Query: 194 SGESYGGHYIPQLAELIFDRNKDKNKYPFINLKGFIVGNPETE 236
+GESY G Y+P +A +FD+N + +NL G +G+ T+
Sbjct: 554 TGESYAGKYVPAIAHYVFDKNTQLEESERLNLAGVAIGDGLTD 682
>TC17005 similar to UP|CP22_HORVU (P55748) Serine carboxypeptidase II-2
precursor (CP-MII.2) (Fragment) , partial (28%)
Length = 564
Score = 128 bits (322), Expect = 2e-30
Identities = 60/117 (51%), Positives = 80/117 (68%)
Frame = +2
Query: 369 WEVCNNSILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPL 428
WE C+ I + S ++L ++ +LI GL+IW++SG+ D +PV TRY ++AL LP
Sbjct: 29 WETCSEVINTHWKDSPRTVLNVFQELIHSGLRIWVFSGNTDAVIPVTSTRYSIDALKLPT 208
Query: 429 KSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
S WR WY D +VGG EY GLT+VTVRGAGH VPL++PK AL+L SFLAG +P
Sbjct: 209 VSPWRPWYDDEEVGGWTQEYAGLTFVTVRGAGHEVPLHRPKLALTLFKSFLAGTSMP 379
>TC8804 similar to UP|Q8L6A7 (Q8L6A7) Carboxypeptidase type III, partial
(63%)
Length = 1210
Score = 115 bits (288), Expect = 2e-26
Identities = 95/358 (26%), Positives = 151/358 (41%), Gaps = 22/358 (6%)
Frame = +3
Query: 146 PVGVGFSYTNTSSDLTILEDNFVAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQ 205
P G GFSY+ D+ E V+ D Y+FL + P++ S FFI+GESY GHYIP
Sbjct: 3 PTGTGFSYSTDKRDIRHDEAG-VSNDLYDFLQAFFTEHPEYASNDFFITGESYAGHYIPA 179
Query: 206 LAELIFDRNKDKNKYPFINLKGFIVGNPETEDYYDYKGLLEYAWSHAVISDQQYDKAKQV 265
A + NK K INLKGF +GN T+ + YK +YA +I YD+ +V
Sbjct: 180 FAARVHKGNKAKEGI-HINLKGFAIGNGLTDPGFQYKAYPDYAMDMGIIKQADYDRINKV 356
Query: 266 ----CDFKQFQWSNE-----------CNKAMNEVFQDYSEIDIYNIYAPKCRLNSTSAIA 310
C+ + CN N + +I+ Y+I
Sbjct: 357 MVPACEVAIKLCGTDGKIACTAAYFVCNTIFNSIMSHAGDINYYDI-------------- 494
Query: 311 SEGLEQLTKGRNDYRMKRKRIFGGYDPCYS-TYAEKYFNRIDVQSSFHVNTERGNTNITW 369
RK+ G CY + EK+ N+ V+ + V +I +
Sbjct: 495 -----------------RKKCEGSL--CYDFSNMEKFLNQRSVRDALGVG------DIDF 599
Query: 370 EVCNNSILQTYNFSVFSILPI-YTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLP- 427
C+ ++ Q L + L++ G+ + IY+G+ D +G V A+
Sbjct: 600 VSCSTTVYQAMLVDWMRDLEVGIPTLLEDGINLLIYAGEYDLICNWLGNSRWVHAMQWSG 779
Query: 428 ----LKSSWRSWYLDNQVGGRIVEYEGLTYVTVRGAGHLVPLNKPKEALSLIHSFLAG 481
+ S + +D G + Y L+++ VR AGH+VP+++PK AL ++ + G
Sbjct: 780 QKEFVASPEVPFTVDESEAGLLKNYGPLSFLKVRDAGHMVPMDQPKAALEMLKRWTQG 953
>BP039437
Length = 524
Score = 108 bits (269), Expect = 3e-24
Identities = 52/107 (48%), Positives = 71/107 (65%), Gaps = 1/107 (0%)
Frame = -2
Query: 381 NFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQ 440
N S ++LPI +L G+++WI+SGD DGRVPV T+Y ++ + LP+K+ WR W+ +
Sbjct: 523 NDSPSTVLPILHELFSNGIRVWIFSGDTDGRVPVTSTKYSIKKMNLPIKNVWRPWFAYGE 344
Query: 441 VGGRIVEYEG-LTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLPT 486
VGG Y+G LT+ TVR AGH VP +P AL LI SFL G LP+
Sbjct: 343 VGGYTEVYKGDLTFATVREAGHQVPSYQPARALVLIKSFLDGTPLPS 203
>TC13125 similar to PIR|C96759|C96759 protein serine carboxypeptidase
T18K17.3 [imported] - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (29%)
Length = 698
Score = 74.7 bits (182), Expect(3) = 2e-19
Identities = 49/153 (32%), Positives = 82/153 (53%), Gaps = 6/153 (3%)
Frame = +1
Query: 110 GAAIEIGPLLVN-KNGEG----LNFNPYSWNQEANLLFVESPVGVGFSYTNTSSDLTILE 164
G A +IGPL K +G L SW + ++++V+ P+G GFSY + ++T
Sbjct: 253 GLAFQIGPLKFEIKEYDGSVPKLVLRSQSWTKLCSIMYVDLPLGTGFSY---AKNVTAQR 423
Query: 165 DNF-VAEDAYNFLVNWLQRFPQFKSRGFFISGESYGGHYIPQLAELIFDRNKDKNKYPFI 223
++ + A+ FL WL P+F S F++ +SY G P + + I + N +K P+I
Sbjct: 424 SDWKLVHHAHQFLRKWLIDHPEFLSNEFYMGADSYSGIPAPAIVQEISNGN-EKGLQPWI 600
Query: 224 NLKGFIVGNPETEDYYDYKGLLEYAWSHAVISD 256
NL+G+++GNP T Y + + +A +ISD
Sbjct: 601 NLQGYLLGNPIT-TYRELNYQIPFAHGMGLISD 696
Score = 30.0 bits (66), Expect(3) = 2e-19
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +3
Query: 94 LLLWLNGGPGCSS 106
L+LWL GGPGCSS
Sbjct: 207 LVLWLTGGPGCSS 245
Score = 27.3 bits (59), Expect(3) = 2e-19
Identities = 9/24 (37%), Positives = 16/24 (66%)
Frame = +2
Query: 71 NEDHGRALFYWFFEAQSEPSKKPL 94
N+D +FY+F ++++ P K PL
Sbjct: 137 NDDDDMQVFYYFIKSENNPQKDPL 208
>BP057139
Length = 545
Score = 86.7 bits (213), Expect = 8e-18
Identities = 48/122 (39%), Positives = 66/122 (53%), Gaps = 1/122 (0%)
Frame = -1
Query: 361 ERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYC 420
ER + WE+C + I +N S++P + L K G + IYSGD D VP G+
Sbjct: 545 ERDSVXGPWELCTSRIKYYHNTGE-SMIPYHKNLTKLGYRALIYSGDHDMCVPFTGSEAW 369
Query: 421 VEALGLPLKSSWRSWYLDNQVGGRIVEYE-GLTYVTVRGAGHLVPLNKPKEALSLIHSFL 479
+LG + WR W +NQV G + YE L ++TV+GAGH VP KP+EAL +L
Sbjct: 368 TRSLGYKIVDEWRQWRSNNQVAGYLQAYENNLIFLTVKGAGHTVPEYKPREALDFYRRWL 189
Query: 480 AG 481
G
Sbjct: 188 EG 183
>TC17210 similar to UP|Q9FH05 (Q9FH05) Serine carboxypeptidase II-like
(AT5g42240/K5J14_4), partial (24%)
Length = 565
Score = 84.0 bits (206), Expect = 5e-17
Identities = 41/105 (39%), Positives = 63/105 (59%), Gaps = 5/105 (4%)
Frame = +2
Query: 386 SILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSS----WRSWYLDNQV 441
+ILPI +++K + +W++SGD D VP++G+R + L L+ + +W+ QV
Sbjct: 11 NILPILKRIVKNHIPVWVFSGDQDSVVPLLGSRTLIRELAHELQYKVTVPYGAWFHKGQV 190
Query: 442 GGRIVEYEG-LTYVTVRGAGHLVPLNKPKEALSLIHSFLAGDRLP 485
GG + EY LT+ VRGA H+VP +P AL L +F+ G RLP
Sbjct: 191 GGWVTEYGNLLTFAPVRGAAHMVPYAQPSRALGLFSAFVRGSRLP 325
>BP031317
Length = 124
Score = 83.2 bits (204), Expect = 9e-17
Identities = 40/41 (97%), Positives = 40/41 (97%)
Frame = +2
Query: 13 MNMNVILSVHFLCFFLLTTSFIKTYGINVGANESDRIVDLP 53
MNMNV LSVHFLCFFLLTTSFIKTYGINVGANESDRIVDLP
Sbjct: 2 MNMNVDLSVHFLCFFLLTTSFIKTYGINVGANESDRIVDLP 124
>TC9695 similar to UP|Q8L7B2 (Q8L7B2) Serine carboxypeptidase 1-like
protein (Serine carboxypeptidase 1 precursor-like
protein), partial (29%)
Length = 597
Score = 81.6 bits (200), Expect = 3e-16
Identities = 48/147 (32%), Positives = 73/147 (49%), Gaps = 2/147 (1%)
Frame = +1
Query: 337 PCYSTYAEK-YFNRIDVQSSFHVNTERGNTNITWEVCNNSILQTYNFSVFSILPIYTKLI 395
PC K + N V+ + H T + + W++C I Y+ S++ + L
Sbjct: 4 PCTDDVVAKLWLNNEAVRKAIH--TAKTSLVSQWDLCTGRI--RYHHDAGSMIKYHKHLT 171
Query: 396 KGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSSWRSWYLDNQVGGRIVEYE-GLTYV 454
G + IYSGD D VP G+ ++G + WR W+ ++QV G Y+ LT++
Sbjct: 172 SQGYRALIYSGDHDMCVPFTGSEAWTRSMGYKIVDEWRPWFSNDQVAGFTQGYDKNLTFM 351
Query: 455 TVRGAGHLVPLNKPKEALSLIHSFLAG 481
T++GAGH VP KP+EAL FL G
Sbjct: 352 TIKGAGHTVPEYKPREALEFYTHFLTG 432
>TC17887 weakly similar to UP|AAQ91191 (AAQ91191)
1-O-sinapoylglucose:choline sinapoyltransferase, partial
(11%)
Length = 595
Score = 75.5 bits (184), Expect = 2e-14
Identities = 37/114 (32%), Positives = 63/114 (54%), Gaps = 5/114 (4%)
Frame = +1
Query: 372 CNNSILQTYNFSVFSILPIYTKLIKGGLKIWIYSGDADGRVPVIGTRYCVEALGLPLKSS 431
CN ++ T S +++ Y L K L+ +Y D D VP +GT++ +++L + +
Sbjct: 100 CNKTLAYTTTLS--NVVESYRNLTKANLQALVYCSDLDMSVPQLGTQHWIKSLNMSISDK 273
Query: 432 WRSWYLDNQVGG-----RIVEYEGLTYVTVRGAGHLVPLNKPKEALSLIHSFLA 480
WR+W+++ QV G ++ E TYV V+GAGH+ KPKE +I S+ +
Sbjct: 274 WRAWFVEGQVAGFTEVYKMKEDHYFTYVAVKGAGHVAQTFKPKEVYHVIKSWFS 435
>BP044220
Length = 386
Score = 68.9 bits (167), Expect = 2e-12
Identities = 40/105 (38%), Positives = 53/105 (50%), Gaps = 1/105 (0%)
Frame = +1
Query: 42 GANESDRIVDLPGVPSSPSVSHFSGYITVNE-DHGRALFYWFFEAQSEPSKKPLLLWLNG 100
G D +V LP P S + I ++ + + W S P WLNG
Sbjct: 76 GYPAEDLVVKLPVNPMLGSGNMLDMLILMSSMERACSTILWRLNMTMATSPSPF--WLNG 249
Query: 101 GPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLLFVES 145
GPGCSS+G GA E+GP + +G GL N +WN+ +NLLFVES
Sbjct: 250 GPGCSSIGGGAFTELGPFFPSGDGRGLRRNSKAWNKASNLLFVES 384
>TC15919 similar to UP|Q9XH61 (Q9XH61) Serine carboxypeptidase, partial
(21%)
Length = 523
Score = 67.0 bits (162), Expect = 7e-12
Identities = 38/106 (35%), Positives = 58/106 (53%), Gaps = 7/106 (6%)
Frame = +2
Query: 43 ANESDRIVDLP-------GVPSSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLL 95
+N + RIV+ P G S ++H +GY + H +FY+FFE+++ K P++
Sbjct: 215 SNNTTRIVEKPLRFPNVLGGVSLDDLAHRAGYYPIQHSHAARMFYFFFESRNS-KKDPVV 391
Query: 96 LWLNGGPGCSSVGYGAAIEIGPLLVNKNGEGLNFNPYSWNQEANLL 141
+WL GGPGCSS E GP + N L +N Y W++ +NLL
Sbjct: 392 IWLTGGPGCSS-ELAVFYENGPFKIADN-MSLVWNDYGWDKVSNLL 523
>TC17530 similar to UP|Q8L6A7 (Q8L6A7) Carboxypeptidase type III, partial
(20%)
Length = 538
Score = 64.7 bits (156), Expect = 3e-11
Identities = 32/88 (36%), Positives = 53/88 (59%)
Frame = +1
Query: 57 SSPSVSHFSGYITVNEDHGRALFYWFFEAQSEPSKKPLLLWLNGGPGCSSVGYGAAIEIG 116
S+ + H++GY + H +FY+FFE+++ + P+++WL GGPGCSS E G
Sbjct: 280 SADDLGHYAGYYPIQHSHAARMFYFFFESRNS-QEDPVVIWLTGGPGCSS-ELA*FNENG 453
Query: 117 PLLVNKNGEGLNFNPYSWNQEANLLFVE 144
P + N L +N Y W++ +NLL+V+
Sbjct: 454 PFKIADN-LTLVWNEYGWDKVSNLLYVD 534
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.139 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,576,554
Number of Sequences: 28460
Number of extensions: 144774
Number of successful extensions: 728
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 698
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 699
length of query: 488
length of database: 4,897,600
effective HSP length: 94
effective length of query: 394
effective length of database: 2,222,360
effective search space: 875609840
effective search space used: 875609840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0333b.9


