

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0333b.12
(327 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
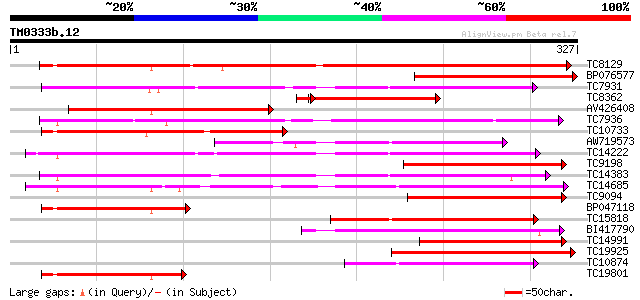
Score E
Sequences producing significant alignments: (bits) Value
TC8129 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (90%) 281 1e-76
BP076577 196 3e-51
TC7931 weakly similar to UP|Q43640 (Q43640) Dioxygenase, partial... 148 1e-36
TC8362 similar to UP|AAQ93035 (AAQ93035) Gibberellin 2-oxidase, ... 139 4e-36
AV426408 136 5e-33
TC7936 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, pa... 120 2e-28
TC10733 similar to UP|Q9SQ81 (Q9SQ81) Gibberellin 2 beta-hydroxy... 119 5e-28
AW719573 110 3e-25
TC14222 similar to UP|Q41681 (Q41681) 1-aminocyclopropane-1-carb... 104 2e-23
TC9198 similar to UP|Q9SQ80 (Q9SQ80) Gibberellin 2 beta-hydroxyl... 102 1e-22
TC14383 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-ca... 102 1e-22
TC14685 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partia... 97 3e-21
TC9094 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (28%) 96 1e-20
BP047118 95 2e-20
TC15818 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxyge... 92 9e-20
BI417790 92 2e-19
TC14991 similar to UP|O04162 (O04162) Dioxygenase, partial (28%) 89 8e-19
TC19925 weakly similar to UP|Q8GXN0 (Q8GXN0) SRG1 like protein, ... 86 6e-18
TC10874 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxyge... 85 1e-17
TC19801 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (34%) 83 5e-17
>TC8129 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (90%)
Length = 1278
Score = 281 bits (718), Expect = 1e-76
Identities = 148/313 (47%), Positives = 205/313 (65%), Gaps = 6/313 (1%)
Frame = +2
Query: 18 DLPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQ 77
++P VDL E LIV A +E+GFFK+INH I E I+ +E FF +P +K++
Sbjct: 194 EVPEVDLS--HPEAKTLIVNACKEFGFFKIINHDIPMELISNLENEALSFFMQPQSEKEK 367
Query: 78 AAP----GYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSN--FRSTVSAYTE 131
A+ GYG K IG NGD+G VEYLL N N P + + + ++ + F S V+ YT
Sbjct: 368 ASLDDPFGYGSKRIGKNGDVGWVEYLLFNTN-PDVISPNPLLLLEQNQNVFSSAVNDYTL 544
Query: 132 AVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSYN 191
AV+ L+CEILE+MA+GLG++ VFSRLIRD +SD + RLNHYP E ++
Sbjct: 545 AVKNLSCEILELMADGLGIEQRNVFSRLIRDEKSDCIFRLNHYPAC---GELELQAIGGR 715
Query: 192 NKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRF 251
N +GFGEH+DPQ++++LRSN+ SGLQI L+DG W+ + PD +F +NVGD L+VMTNG F
Sbjct: 716 NLIGFGEHTDPQMISVLRSNNTSGLQICLRDGTWVSIPPDQTSFFINVGDSLQVMTNGSF 895
Query: 252 VSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYKKATYSL 311
SV+HR ++++ SR+SM YFG PL + P LV+ E SL++ FTW +YKKA Y L
Sbjct: 896 RSVKHRVLSDTAMSRVSMIYFGGAPLTEKLAPLPSLVSKEEDSLYKEFTWGEYKKAAYKL 1075
Query: 312 RLGDSRMELFRNN 324
RL D+R+ F +
Sbjct: 1076RLADNRLTPFEKS 1114
>BP076577
Length = 392
Score = 196 bits (499), Expect = 3e-51
Identities = 94/94 (100%), Positives = 94/94 (100%)
Frame = -2
Query: 234 AFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERP 293
AFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERP
Sbjct: 391 AFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERP 212
Query: 294 SLFRPFTWADYKKATYSLRLGDSRMELFRNNNDM 327
SLFRPFTWADYKKATYSLRLGDSRMELFRNNNDM
Sbjct: 211 SLFRPFTWADYKKATYSLRLGDSRMELFRNNNDM 110
>TC7931 weakly similar to UP|Q43640 (Q43640) Dioxygenase, partial (29%)
Length = 1477
Score = 148 bits (374), Expect = 1e-36
Identities = 99/296 (33%), Positives = 154/296 (51%), Gaps = 10/296 (3%)
Frame = +1
Query: 19 LPIVDLKA-EKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQ 77
+P+VDL +++E + I KASEEYGFF+VINHG+S+E I F A P +K
Sbjct: 133 VPVVDLGGHDRAETLKQIFKASEEYGFFQVINHGVSNELIEDTLNIFKEFHAMPAEEKLS 312
Query: 78 AA---PGYGC-----KNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAY 129
+ P C + I + + L + TPS + + + P+ +R V Y
Sbjct: 313 ESSRDPNGSCMLYTSREINNKDCIQFWKDTLRHPCTPS-EEFMEFWPLKPARYREVVEKY 489
Query: 130 TEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPS 189
T+ +REL +ILE++ EGLG+ + L + +L NHYPP P
Sbjct: 490 TKELRELGSQILEMLCEGLGLDPRYCCAGL----SENPLLLSNHYPPC----------PE 627
Query: 190 YNNKVGFGEHSDPQILTIL-RSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTN 248
+ +G +H DP ++TIL + DV+ LQ+ +DG WI ++P P AF VN+G +L++++N
Sbjct: 628 PSLTLGLSKHRDPTLVTILLQDTDVNALQV-FKDGEWISIEPIPYAFVVNIGLLLQIISN 804
Query: 249 GRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADY 304
GR V V HR +TNS SR ++AYF P + LV ++R + ++
Sbjct: 805 GRLVGVEHRVVTNSSISRTTVAYFIRPKKEEIVEPAKALVRTGAHPIYRSIAFEEF 972
>TC8362 similar to UP|AAQ93035 (AAQ93035) Gibberellin 2-oxidase, partial
(20%)
Length = 675
Score = 139 bits (350), Expect(2) = 4e-36
Identities = 67/76 (88%), Positives = 68/76 (89%)
Frame = +2
Query: 173 HYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDP 232
H PI NSNNREDKSP YNNKVGFGEHSDPQILTILRSNDV GLQISLQ+GVWIPVKPDP
Sbjct: 29 HLIPIHNSNNREDKSPPYNNKVGFGEHSDPQILTILRSNDVCGLQISLQEGVWIPVKPDP 208
Query: 233 EAFCVNVGDVLEVMTN 248
EAFCVNVGDVLE N
Sbjct: 209 EAFCVNVGDVLEDNAN 256
Score = 28.5 bits (62), Expect(2) = 4e-36
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = +1
Query: 166 DSVLRLNHYPP 176
DSVLRLNHYPP
Sbjct: 1 DSVLRLNHYPP 33
>AV426408
Length = 385
Score = 136 bits (342), Expect = 5e-33
Identities = 64/122 (52%), Positives = 87/122 (70%), Gaps = 4/122 (3%)
Frame = +3
Query: 35 IVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQAAP----GYGCKNIGFN 90
+V+A EEYGFFKV+NH + E ++++EE G FF+K +K QA P GYGC+NIG N
Sbjct: 9 VVRACEEYGFFKVVNHNVPREVVSRLEEKGTEFFSKSTSEKTQAGPATPFGYGCRNIGLN 188
Query: 91 GDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYTEAVRELACEILEVMAEGLGV 150
GDMG++EYLLL+ N S + SK+++ DP+ F VS Y E +ELAC+ILE++AEGL V
Sbjct: 189 GDMGDLEYLLLHTNPLSFTETSKTIANDPTKFSCAVSDYIEEAKELACKILELVAEGLWV 368
Query: 151 QD 152
D
Sbjct: 369 PD 374
>TC7936 similar to UP|Q9SP53 (Q9SP53) Flavanone 3-hydroxylase, partial
(94%)
Length = 1492
Score = 120 bits (302), Expect = 2e-28
Identities = 88/316 (27%), Positives = 155/316 (48%), Gaps = 14/316 (4%)
Frame = +3
Query: 18 DLPIVDLKA------EKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKP 71
++P++ L +SE+ IV+A E +G F+V++HG+ E ++ M FFA P
Sbjct: 258 EIPVISLAGIDEVDDRRSEICNKIVEACENWGIFQVVDHGVDTELVSHMTTLAKEFFALP 437
Query: 72 MPQKKQAAPGYGCKNIGF-------NGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRS 124
P++K G K GF + + ++ + P + P+ +++
Sbjct: 438 -PEEKLRFDMTGGKKGGFIVSSHLQGESVQDWREIVTYFSYPIRNRDYSRWPDTPAGWKA 614
Query: 125 TVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNRE 184
Y+E + LAC++LEV++E +G++ + ++ D + +N+YP
Sbjct: 615 VTEEYSEKLMGLACKLLEVLSEAMGLEKEALTKAC---VDMDQKVVVNYYP--------- 758
Query: 185 DKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDG-VWIPVKPDPEAFCVNVGDVL 243
K P + +G H+DP +T+L + V GLQ + +G WI V+P AF VN+GD
Sbjct: 759 -KCPQPDLTLGLKRHTDPGTITLLLQDQVGGLQATRDNGKTWITVQPVEGAFVVNLGDHG 935
Query: 244 EVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWAD 303
++NGRF + H+A+ NS SR+S+A F P A V P + E+ + P T+A+
Sbjct: 936 HYLSNGRFKNADHQAVVNSNSSRLSIATFQNPAPDA-TVYPLKVREGEKSVMEEPITFAE 1112
Query: 304 YKKATYSLRLGDSRME 319
+ S + +RM+
Sbjct: 1113MYRRKMSKDIELARMK 1160
>TC10733 similar to UP|Q9SQ81 (Q9SQ81) Gibberellin 2 beta-hydroxylase,
partial (52%)
Length = 616
Score = 119 bits (299), Expect = 5e-28
Identities = 68/146 (46%), Positives = 92/146 (62%), Gaps = 4/146 (2%)
Frame = +1
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQ- 77
+P+VDL K + +IVKA EE+GFFKVINHG+S E I+++E FF+ P+ +K++
Sbjct: 190 IPVVDLS--KPDAKSIIVKACEEFGFFKVINHGVSMEAISQLESEALNFFSMPVTEKEKT 363
Query: 78 --AAP-GYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYTEAVR 134
A P GYG K IG NGD+G VEYLLL N + S +P FR + Y AVR
Sbjct: 364 GLANPFGYGNKRIGHNGDVGWVEYLLLKTNQEVNVPVH---SQNPEKFRCVLDDYLCAVR 534
Query: 135 ELACEILEVMAEGLGVQDTLVFSRLI 160
+ CEIL++MAEGL +Q V S+L+
Sbjct: 535 NMGCEILDLMAEGLKIQPKNVLSKLL 612
>AW719573
Length = 521
Score = 110 bits (275), Expect = 3e-25
Identities = 64/172 (37%), Positives = 94/172 (54%), Gaps = 3/172 (1%)
Frame = +2
Query: 119 PSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDI--ESDSVLRLNHYPP 176
P FR + Y + L+ +I+E+MA L + + I +I E +R+N+YPP
Sbjct: 53 PQPFRENLEIYCAELNNLSIKIMELMANALKIDP-----KEITEIYNEGSQTIRMNYYPP 217
Query: 177 ILNSNNREDKSPSYNNKVGFGEHSDPQILTIL-RSNDVSGLQISLQDGVWIPVKPDPEAF 235
P +G H+D LTIL ++ND+ GLQI +DG W+PV+P P AF
Sbjct: 218 C----------PQPERVIGLKSHTDGAGLTILLQANDIEGLQIR-KDGHWVPVQPLPNAF 364
Query: 236 CVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVL 287
+N+GD+LE+ TNG + S+ HRA NS K R+S+A F P + A + P L
Sbjct: 365 IINIGDMLEITTNGIYASIEHRATVNSEKERISLATFYGPNMQATLAPAPSL 520
>TC14222 similar to UP|Q41681 (Q41681) 1-aminocyclopropane-1-carboxylate
oxidase homolog (ACC oxidase) , partial (97%)
Length = 1597
Score = 104 bits (260), Expect = 2e-23
Identities = 88/304 (28%), Positives = 147/304 (47%), Gaps = 7/304 (2%)
Frame = +1
Query: 10 RSEKIVPGDLPIVDLKA----EKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGF 65
R EK + + P+VD+ E+ LI A E +GFF+++NH IS E + +E
Sbjct: 31 REEKKM-ANFPVVDMSKLNTEERVATMELIKDACENWGFFELVNHEISTEFMDTVERLTK 207
Query: 66 GFFAKPMPQK-KQAAPGYGCKNIGFNGDMGEVEYLLLNANTPSIAQISKSVSIDPSNFRS 124
+ + M Q+ K+ G + + D + E + PS + IS+ +D ++R
Sbjct: 208 EHYKRFMEQRFKEMVATKGLETVQSEIDDLDWESTFFLRHLPS-SNISEVPDLD-EDYRK 381
Query: 125 TVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNRE 184
+ + + +LA +L+++ E LG++ + + ++++YPP
Sbjct: 382 VMKEFAVKLEKLAENLLDLLCENLGLEKGYLKKVFYGSKGPNFGTKVSNYPPC------- 540
Query: 185 DKSPSYNNKVGFGEHSDPQ-ILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVL 243
P + G H+D I+ + + + VSGLQ+ L+D WI V P + VN+GD L
Sbjct: 541 ---PKPDLIKGLRAHTDAGGIILLFQDDKVSGLQL-LKDDQWIDVPPMRHSIVVNLGDQL 708
Query: 244 EVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRP-FTWA 302
EV+TNG++ SV HR + + +RMS+A F P A I P LV + S P F +
Sbjct: 709 EVITNGKYKSVMHRVIAQTDGARMSLASFYNPGDDAVISPAPSLVKEDETSQVYPKFVFE 888
Query: 303 DYKK 306
DY K
Sbjct: 889 DYMK 900
>TC9198 similar to UP|Q9SQ80 (Q9SQ80) Gibberellin 2 beta-hydroxylase,
partial (30%)
Length = 555
Score = 102 bits (253), Expect = 1e-22
Identities = 49/94 (52%), Positives = 63/94 (66%)
Frame = +3
Query: 228 VKPDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVL 287
V D ++F + VGD L+VMTNGRF SVRHR + N FKSR+SM YFG PPL I P L
Sbjct: 6 VPSDHDSFFIYVGDSLQVMTNGRFRSVRHRVLANGFKSRLSMIYFGGPPLSQKIAPLPCL 185
Query: 288 VTPERPSLFRPFTWADYKKATYSLRLGDSRMELF 321
+ SL++ FTW +YKK+ Y+ RL D+R+ F
Sbjct: 186 MKGNESSLYKEFTWYEYKKSAYASRLADNRLGNF 287
>TC14383 homologue to UP|Q8W3Y3 (Q8W3Y3) 1-aminocyclopropane-1-carboxylic
acid oxidase, complete
Length = 1249
Score = 102 bits (253), Expect = 1e-22
Identities = 82/306 (26%), Positives = 145/306 (46%), Gaps = 11/306 (3%)
Frame = +3
Query: 18 DLPIVDLKA----EKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMP 73
+ P++ L+ E+ ++ I A E +GFF+++NHGI H+ + +E + M
Sbjct: 45 NFPVISLEKLNGEERKDIKLQIKDACENWGFFELVNHGIPHDVMDTVERLTKEHYRICME 224
Query: 74 QK-KQAAPGYGCKNIGFN-GDMG-EVEYLLLNANTPSIAQISKSVSIDPSNFRSTVSAYT 130
Q+ K+ G + + DM E + L + +I++I +R + +
Sbjct: 225 QRFKELMASKGLEAVQTEVKDMDWESTFHLRHLPESNISEIPDLTD----EYRKVMKDFA 392
Query: 131 EAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPILNSNNREDKSPSY 190
+ +L+ ++L+++ E LG++ + ++ +YPP P
Sbjct: 393 LRIEKLSEDLLDLLCENLGLEKGYLKKAFHGSRGPTFGTKVANYPPC----------PKP 542
Query: 191 NNKVGFGEHSDPQ-ILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNG 249
N G H+D I+ + + + VSGLQ+ L+DG W+ V P + VN+GD LEV+TNG
Sbjct: 543 NLVKGLRAHTDAGGIILLFQDDKVSGLQL-LKDGEWVDVPPMRHSIVVNLGDQLEVITNG 719
Query: 250 RFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLV---TPERPSLFRPFTWADYKK 306
++ SV HR + + +RMS+A F P A I P L+ E+ L+ F + DY K
Sbjct: 720 KYKSVEHRVIAQTDGTRMSIASFYNPGSDAVIYPAPELLEKEAEEKNQLYPKFVFEDYMK 899
Query: 307 ATYSLR 312
L+
Sbjct: 900 LYAGLK 917
>TC14685 weakly similar to UP|Q43383 (Q43383) 2A6 protein, partial (38%)
Length = 1323
Score = 97.4 bits (241), Expect = 3e-21
Identities = 87/328 (26%), Positives = 149/328 (44%), Gaps = 15/328 (4%)
Frame = +1
Query: 10 RSEKIVPGDLPIVDLKA---EKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFG 66
R E ++P +DL A ++ V I +A+ GFF+V+NHG+ + + + A
Sbjct: 271 RPESDSKPEIPTIDLAAVNESRAAVVDQIRRAASSVGFFQVVNHGVPLDLLRRTVAAIKA 450
Query: 67 FFAKPMPQKKQAAP---GYGCKNIGFNGDMGEV------EYLLLNANTPSIAQISKSVSI 117
F +P ++ + G G I N D+ + + L L ++ Q S+
Sbjct: 451 FHEQPEEERARVYTREMGTGVSYIS-NVDLFQSKAASWRDTLQLRMGPTAVDQ-----SM 612
Query: 118 DPSNFRSTVSAYTEAVRELACEILEVMAEGLGVQDTLVFSRLIRDIESDSVLRLNHYPPI 177
P R V + + V + + +++EGLG+ RL + + HY P
Sbjct: 613 IPEVCRQEVMEWDKEVVRVGEVLYGLLSEGLGLDA----GRLSEMGLVQGRVMVGHYYPF 780
Query: 178 LNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQDGVWIPVKPDPEAFCV 237
P + VG H+DP LT++ + + GLQ+ +G W+ VKP A +
Sbjct: 781 C---------PQPDLTVGLNSHADPGALTVVLQDHIGGLQVRTTEG-WVDVKPLDGALVI 930
Query: 238 NVGDVLEVMTNGRFVSVRHRAMTNSF-KSRMSMAYFGAPPLHACIVAP-PVLVTPERPSL 295
N+GD+L++++N + S HR + NS + R+S+A F P + P P L + E+P+L
Sbjct: 931 NIGDLLQIISNEEYKSADHRVLANSANEPRVSVAVFLNPGNREKLFGPLPELTSAEKPAL 1110
Query: 296 FRPFTWADYKKATYSLRL-GDSRMELFR 322
+R FT ++ + L G S FR
Sbjct: 1111YRNFTLDEFLTRFFKKELDGKSLTNYFR 1194
>TC9094 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (28%)
Length = 677
Score = 95.5 bits (236), Expect = 1e-20
Identities = 47/92 (51%), Positives = 62/92 (67%)
Frame = +1
Query: 230 PDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVT 289
PD +F + VGD L+VMTNGRF SV+HR M ++ KSR+SM YFG L I L++
Sbjct: 1 PDHTSFFIYVGDTLQVMTNGRFKSVKHRVMADTTKSRLSMIYFGGASLSEKIAPLESLMS 180
Query: 290 PERPSLFRPFTWADYKKATYSLRLGDSRMELF 321
SL++ FTW++YKKA YS RLGD+R+ F
Sbjct: 181 KGEQSLYQEFTWSEYKKAAYSSRLGDNRLGPF 276
>BP047118
Length = 476
Score = 94.7 bits (234), Expect = 2e-20
Identities = 50/90 (55%), Positives = 62/90 (68%), Gaps = 4/90 (4%)
Frame = -2
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
+P+VDL K + LIVKA EE+GFFKVINHG+ ETI+ +E FF+ P+ QK++A
Sbjct: 298 IPVVDLS--KPDAKTLIVKACEEFGFFKVINHGVPMETISMLESEASKFFSMPLNQKEKA 125
Query: 79 AP----GYGCKNIGFNGDMGEVEYLLLNAN 104
P GYG KNIG NGD+G VEYLLL N
Sbjct: 124 GPPNPFGYGTKNIGQNGDVGWVEYLLLTNN 35
>TC15818 similar to UP|Q9FFF6 (Q9FFF6) Leucoanthocyanidin dioxygenase-like
protein (AT5g05600/MOP10_14), partial (37%)
Length = 618
Score = 92.4 bits (228), Expect = 9e-20
Identities = 51/121 (42%), Positives = 73/121 (60%), Gaps = 1/121 (0%)
Frame = +2
Query: 186 KSPSYNNKVGFGEHSDPQILTILRSND-VSGLQISLQDGVWIPVKPDPEAFCVNVGDVLE 244
K P + +G HSDP LTIL +D VSGLQ+ + WI VKP P AF +N+GD ++
Sbjct: 8 KCPQPDLTLGLSPHSDPGGLTILLPDDFVSGLQVR-KGNYWITVKPVPNAFIINIGDQIQ 184
Query: 245 VMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADY 304
V++N + SV HR + NS + R+S+A F P I LVT E+P+L+ P T+ +Y
Sbjct: 185 VLSNTIYKSVEHRVIVNSNQDRVSLAMFYNPKSDLLIEPAKELVTEEKPALYPPMTYDEY 364
Query: 305 K 305
+
Sbjct: 365 R 367
>BI417790
Length = 610
Score = 91.7 bits (226), Expect = 2e-19
Identities = 50/156 (32%), Positives = 83/156 (53%), Gaps = 4/156 (2%)
Frame = +3
Query: 169 LRLNHYPPILNSNNREDKSPSYNNKVGFGEHSDPQILTILRSNDVSGLQISLQ-DGVWIP 227
+RLN+YPP P+ + +G G H D LTIL ++VSGLQ+ + DG W+
Sbjct: 6 IRLNYYPPC----------PNPDIVLGCGRHKDSGALTILAQDEVSGLQVRRKCDGQWVR 155
Query: 228 VKPDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVL 287
+ P P+A+ +NVGDV++V N + SV HR N K+R+S +F P + + L
Sbjct: 156 LNPLPDAYIINVGDVIQVWCNDAYESVEHRVKLNPEKARLSYPFFLYPSHYTNVEPLEEL 335
Query: 288 VTPERPSLFRPFTWADY---KKATYSLRLGDSRMEL 320
E P + P+ W + +K + ++L + +++
Sbjct: 336 TNEENPPKYTPYNWGKFFVTRKRSNFMKLDEENVQV 443
>TC14991 similar to UP|O04162 (O04162) Dioxygenase, partial (28%)
Length = 507
Score = 89.4 bits (220), Expect = 8e-19
Identities = 43/86 (50%), Positives = 59/86 (68%), Gaps = 1/86 (1%)
Frame = +2
Query: 237 VNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVT-PERPSL 295
+NVGD L+VMTNGRF SV+HR + N F+SR+SM YFG PPL I P L+ + SL
Sbjct: 5 INVGDSLQVMTNGRFRSVKHRVLANGFQSRLSMIYFGGPPLSEKIAPLPSLMKGGKEESL 184
Query: 296 FRPFTWADYKKATYSLRLGDSRMELF 321
++ FTW +Y+ ++Y RL D+R+ F
Sbjct: 185 YKEFTWFEYQNSSYGSRLADNRLGHF 262
>TC19925 weakly similar to UP|Q8GXN0 (Q8GXN0) SRG1 like protein, partial
(19%)
Length = 613
Score = 86.3 bits (212), Expect = 6e-18
Identities = 43/107 (40%), Positives = 66/107 (61%), Gaps = 1/107 (0%)
Frame = +1
Query: 221 QDGVWIPVKPDPEAFCVNVGDVLEVMTNGRFVSVRHRAMTNSFKSRMSMAYFGAPPLHAC 280
+DG W+PV P AF ++VGD++E++T+G + S+ HRA NS K R+S+A F P ++
Sbjct: 10 KDGKWVPVTPLSHAFVIHVGDIMEILTHGIYRSIEHRATINSEKERISLASFHRPLMNQV 189
Query: 281 IVAPPVLVTPERPSLFRPFTWADYKKATYSLRL-GDSRMELFRNNND 326
I P LVTPERP+LF+ D+ + +S L G + + R N+
Sbjct: 190 IGPTPSLVTPERPALFKTIAVEDFYRVFFSRPLKGKTLLNAMRIENE 330
>TC10874 similar to UP|Q8LF06 (Q8LF06) Leucoanthocyanidin dioxygenase-like
protein, partial (38%)
Length = 723
Score = 85.1 bits (209), Expect = 1e-17
Identities = 46/113 (40%), Positives = 68/113 (59%), Gaps = 1/113 (0%)
Frame = +2
Query: 194 VGFGEHSDPQILTILRSND-VSGLQISLQDGVWIPVKPDPEAFCVNVGDVLEVMTNGRFV 252
+G HSDP +T+L +D V GLQ+ D WI V P AF VN+GD ++V++N +
Sbjct: 8 LGLSSHSDPGGMTMLLPDDQVRGLQVRKGDD-WITVNPARHAFIVNIGDQIQVLSNAIYT 184
Query: 253 SVRHRAMTNSFKSRMSMAYFGAPPLHACIVAPPVLVTPERPSLFRPFTWADYK 305
SV HR + NS K R+S+A+F P I LVTP++P+L+ T+ +Y+
Sbjct: 185 SVEHRVIVNSDKERVSLAFFYNPKSDIPIEPVKELVTPDKPALYPAMTFDEYR 343
>TC19801 similar to UP|Q9XG83 (Q9XG83) GA 2-oxidase, partial (34%)
Length = 508
Score = 83.2 bits (204), Expect = 5e-17
Identities = 43/88 (48%), Positives = 57/88 (63%), Gaps = 4/88 (4%)
Frame = +2
Query: 19 LPIVDLKAEKSEVTRLIVKASEEYGFFKVINHGISHETIAKMEEAGFGFFAKPMPQKKQA 78
+P+VDL E LIV+A +E+GFFK+INHG+ + ++ +E FF KP +K +A
Sbjct: 251 IPVVDLS--DPEAKSLIVQACKEFGFFKLINHGVPLDLMSNLENEAVRFFRKPQTEKDRA 424
Query: 79 AP----GYGCKNIGFNGDMGEVEYLLLN 102
P GYG K IG NGD+G VEYLLLN
Sbjct: 425 GPPDPFGYGSKKIGSNGDVGWVEYLLLN 508
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,022,793
Number of Sequences: 28460
Number of extensions: 65040
Number of successful extensions: 450
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 414
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 420
length of query: 327
length of database: 4,897,600
effective HSP length: 91
effective length of query: 236
effective length of database: 2,307,740
effective search space: 544626640
effective search space used: 544626640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0333b.12


