

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
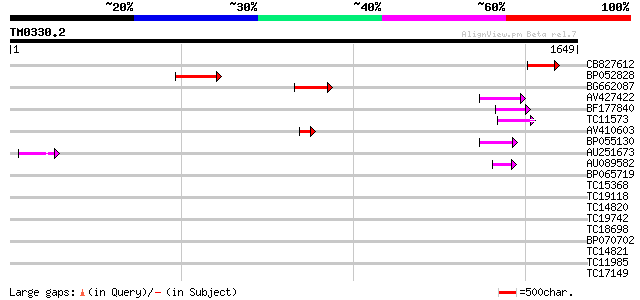
Query= TM0330.2
(1649 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB827612 118 9e-27
BP052828 115 6e-26
BG662087 97 3e-20
AV427422 72 9e-13
BF177840 62 7e-10
TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase regi... 58 1e-08
AV410603 58 1e-08
BP055130 54 2e-07
AU251673 47 3e-05
AU089582 46 4e-05
BP065719 36 0.056
TC15368 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (11%) 33 0.36
TC19118 weakly similar to UP|Q84YE8 (Q84YE8) Expressed protein-l... 33 0.47
TC14820 homologue to UP|HS80_LYCES (P36181) Heat shock cognate p... 32 0.62
TC19742 weakly similar to UP|SDF2_ARATH (Q93ZE8) Stromal cell-de... 32 0.62
TC18698 32 0.81
BP070702 32 1.1
TC14821 homologue to UP|AAR12195 (AAR12195) Molecular chaperone ... 31 1.4
TC11985 similar to GB|AAF40452.1|7211981|F13M7 ESTs gb|N65605, g... 31 1.8
TC17149 similar to GB|AAP37818.1|30725592|BT008459 At1g43860 {Ar... 31 1.8
>CB827612
Length = 277
Score = 118 bits (295), Expect = 9e-27
Identities = 55/92 (59%), Positives = 70/92 (75%)
Frame = +1
Query: 1506 ACHLPVELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHD 1565
ACHLPVE+EH+AYWA++ N + + A +R LQL +L+E RL AYES+ IYKEKTK HD
Sbjct: 1 ACHLPVEIEHRAYWAVKSCNXEMQQAGIERKLQLQQLEELRLGAYESSRIYKEKTKHXHD 180
Query: 1566 RKILNREFVSGQLVLLFNSRLRLFPGKLKSRW 1597
+ I +EF GQ VLLFNSRL+L GKL+S+W
Sbjct: 181 KMISRKEFSVGQQVLLFNSRLKLMAGKLRSKW 276
>BP052828
Length = 523
Score = 115 bits (288), Expect = 6e-26
Identities = 61/138 (44%), Positives = 92/138 (66%), Gaps = 4/138 (2%)
Frame = +2
Query: 483 LCDLGASINLMPLSMFKRLNLGEVTPTMISLQMADRSLKTPYGIIEDVVVKVDKFVFPVD 542
+ DLGA IN+MP S++ L+ G + T + +Q+A+RS P G++EDV+V+V++ +FP D
Sbjct: 107 MLDLGAGINVMPTSIYNILHPGPLQHTGLIVQLANRSNARPKGVVEDVLVQVNELIFPAD 286
Query: 543 FVILDME---EDSKVPLILGRPFLATGEAEIKVAKGTLTLKVGEDEVLFNIFDSLKH-RA 598
F ILDME + S+ P+ILGRPF+ T + +I V GT++++ G+ FNI D++KH
Sbjct: 287 FYILDMEGETKSSRAPIILGRPFMKTAKTKIDVDNGTMSMEFGDIIAKFNIDDAMKHPLE 466
Query: 599 DEEVFRCEVVDELVCEEF 616
D VF VV ELV + F
Sbjct: 467 DHSVFHIGVVSELVDDTF 520
>BG662087
Length = 373
Score = 96.7 bits (239), Expect = 3e-20
Identities = 44/110 (40%), Positives = 69/110 (62%)
Frame = +1
Query: 828 KWRVCIDYRRLNSVTRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKT 887
KWR+ +DY LN KD +PLP ID+++D + ++ +D YSGY+QI + P D++KT
Sbjct: 19 KWRMWVDYTDLNKACPKDSYPLPSIDKLVDGASDNELLSLMDAYSGYHQIKMHPSDEDKT 198
Query: 888 AFTCPYDVFAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSV 937
AF + Y+ +PFGL NA AT+Q M +F D + +E+++D+ V
Sbjct: 199 AFMTARVNYCYQTIPFGLKNAGATYQXLMDRVFXDXVGRNMEVYLDNMIV 348
>AV427422
Length = 417
Score = 71.6 bits (174), Expect = 9e-13
Identities = 41/135 (30%), Positives = 68/135 (50%), Gaps = 1/135 (0%)
Frame = +1
Query: 1365 PPSFGCQYILLAVDYVSKWVEAAALS-TNDSKVVVAFLKKNIFTRFGVPRAIISDGGTHF 1423
P S G + +L+ VD +SK+ L +KV+ + + GVP +I+SD F
Sbjct: 13 PKSKGYEAVLVVVDRLSKFSHFVPLKHPYTAKVIADIFVREVVRLHGVPLSIVSDRDPLF 192
Query: 1424 CNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRILEKVVDSSRKDWSRKLDDAL 1483
+ ++ L + G K K+ST YHP++ GQ E+ NR L+ L + K W+ + A
Sbjct: 193 MSNFWKELFKMQGTKLKMSTAYHPESDGQTEVVNRCLETYLRCFIADQPKSWAHWVPWAE 372
Query: 1484 WAYRTAFKTPIGTSP 1498
+ Y T++ G +P
Sbjct: 373 YWYNTSYHVSTGQTP 417
>BF177840
Length = 410
Score = 62.0 bits (149), Expect = 7e-10
Identities = 34/100 (34%), Positives = 51/100 (51%)
Frame = +2
Query: 1414 AIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRILEKVVDSSRK 1473
+I+SD T F + + +L K G K ST HPQT GQ E+ N+ L +L V++ + K
Sbjct: 14 SIVSDRDTKFISHFWRTLWGKVGTKLLYSTTCHPQTDGQTEVVNKTLSTLLRSVLERNLK 193
Query: 1474 DWSRKLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVEL 1513
W L +AY + SPF +V+G P++L
Sbjct: 194 MWETWLPHIEFAYNRVVHSTTKHSPFEIVYGYNPLTPLDL 313
>TC11573 similar to UP|Q9M6N4 (Q9M6N4) Pol protein integrase region
(Fragment), partial (10%)
Length = 572
Score = 58.2 bits (139), Expect = 1e-08
Identities = 30/111 (27%), Positives = 56/111 (50%)
Frame = +2
Query: 1418 DGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRILEKVVDSSRKDWSR 1477
+ G F ++ + + G++ + S+ HPQT+GQ E +N+ + + +++ + + W
Sbjct: 8 ENGIQFTSKQTQDFCDGMGIQMRFSSVKHPQTNGQTEAANKVILKGIKRRLYEAEGRWID 187
Query: 1478 KLDDALWAYRTAFKTPIGTSPFHLVFGKACHLPVELEHKAYWAIRKLNFDW 1528
+L LW+Y T ++ I +PF L +G LPVE+ NF W
Sbjct: 188 ELPIVLWSYNTMPQSSIKETPF*LTYGADTMLPVEIS----------NFSW 310
>AV410603
Length = 162
Score = 58.2 bits (139), Expect = 1e-08
Identities = 26/48 (54%), Positives = 35/48 (72%)
Frame = +1
Query: 842 TRKDHFPLPFIDQMLDKLAGHQYYCFLDGYSGYNQICVAPEDQEKTAF 889
T KD FP+P +D++LD+L G Q++ LD SGY+QI V PED+ KT F
Sbjct: 4 TVKDSFPMPTVDELLDELRGSQFFSKLDLRSGYHQILVKPEDRHKTVF 147
>BP055130
Length = 567
Score = 53.9 bits (128), Expect = 2e-07
Identities = 36/112 (32%), Positives = 55/112 (48%), Gaps = 2/112 (1%)
Frame = +2
Query: 1366 PSFGCQYILLAVDYVSKWVEAAALSTN--DSKVVVAFLKKNIFTRFGVPRAIISDGGTHF 1423
P G I++ VD +SK+ A N S+V F+ I G+PRAI+SD F
Sbjct: 170 PVCGFTVIIVVVDRLSKYGHFAPHRANYTSSQVAETFVS-TIVKLHGMPRAIVSDRDKAF 346
Query: 1424 CNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRILEKVVDSSRKDW 1475
+ ++ + +G +S+ YHPQT GQ E N+ L+ L V + + W
Sbjct: 347 TSAFWKHFFKLHGTTLNMSSSYHPQTDGQTEALNKCLELYLRCFVHETPRLW 502
>AU251673
Length = 413
Score = 46.6 bits (109), Expect = 3e-05
Identities = 38/127 (29%), Positives = 58/127 (44%), Gaps = 8/127 (6%)
Frame = +2
Query: 26 SEAIRLSLFPFSLKDAAEDWLN----SQPQGSLT-TWEDLAEKFTTRFFPRSLLRKLKND 80
S+ + L F L+ A DW N ++P GS TW D + +F RF P+S+ D
Sbjct: 8 SDTRAVELASFQLEGVARDWYNVLTRAKPVGSPPWTWADFSAEFMNRFLPQSVRDGFVRD 187
Query: 81 IMTFAQSTDENLYEAWEHFKKLLRKCPQHNLTQAECVAKFYDGL---LYSSRFGLDAASS 137
Q+ + E HF L R P + L + E V +F GL L+ S G +++
Sbjct: 188 FERLEQAEGMTVSEYSAHFTHLSRYVP-YPLLEEERVKRFVRGLKEYLFKSVVGSKSSTL 364
Query: 138 GEFDALS 144
E +L+
Sbjct: 365 SEVLSLA 385
>AU089582
Length = 383
Score = 46.2 bits (108), Expect = 4e-05
Identities = 26/68 (38%), Positives = 37/68 (54%)
Frame = +2
Query: 1405 IFTRFGVPRAIISDGGTHFCNRAFESLLEKYGVKHKVSTPYHPQTSGQVEISNRELKRIL 1464
I + GVP +IISD G F + + S G + K+ST +HPQT GQ E + + L+ +L
Sbjct: 44 IVSLHGVPVSIISDRGAQFTSHFWRSFQTALGTRLKMSTAFHPQTDGQSERTIQILEDML 223
Query: 1465 EKVVDSSR 1472
V R
Sbjct: 224 RACVXDLR 247
>BP065719
Length = 567
Score = 35.8 bits (81), Expect = 0.056
Identities = 25/99 (25%), Positives = 44/99 (44%)
Frame = +3
Query: 26 SEAIRLSLFPFSLKDAAEDWLNSQPQGSLTTWEDLAEKFTTRFFPRSLLRKLKNDIMTFA 85
+E +++ FP SL A W + S+ TW L F +FF R + D+ +
Sbjct: 273 NENLKMKYFPSSLTKNAFTWFTTLAPRSVHTWAQLERIFHEQFF-RGECKVSXKDLASVK 449
Query: 86 QSTDENLYEAWEHFKKLLRKCPQHNLTQAECVAKFYDGL 124
+ E++ + F+ L +C H +++ E V GL
Sbjct: 450 RKPAESIDDYLNRFRMLKSRCFTH-VSEHELVVLXAGGL 563
>TC15368 homologue to UP|Q84LL5 (Q84LL5) Cullin 4, partial (11%)
Length = 517
Score = 33.1 bits (74), Expect = 0.36
Identities = 19/83 (22%), Positives = 38/83 (44%)
Frame = +1
Query: 629 IMEGLEIEDQNLERELDSLYHEVNATLSQLECVSSLATKSIWKEELTRDEEIPIEEKSEL 688
+ E +E+ ER ++V+A + ++ + + ++ EL + + PI+
Sbjct: 1 LKETVEVNTSTTERVFQDRQYQVDAAIVRIMKTRKVLSHTLLITELFQQLKFPIKPADLK 180
Query: 689 KPLPSSLKYAYLEEGENKPVILN 711
K + S + YLE +N P I N
Sbjct: 181 KRIESLIDREYLERDKNNPQIYN 249
>TC19118 weakly similar to UP|Q84YE8 (Q84YE8) Expressed protein-like protein,
partial (11%)
Length = 932
Score = 32.7 bits (73), Expect = 0.47
Identities = 28/74 (37%), Positives = 35/74 (46%), Gaps = 5/74 (6%)
Frame = +3
Query: 1438 KHKVSTPYHPQTSGQVEISNR--ELKRILEKVVDS---SRKDWSRKLDDALWAYRTAFKT 1492
+HK+S P HP T + E SNR E +L + DS + S LD+AL R
Sbjct: 3 EHKISDPQHPSTY-KSEQSNRLKEDSMLLMRGFDSVAHTLSLMSSNLDNALQGARELANP 179
Query: 1493 PIGTSPFHLVFGKA 1506
P T FH F KA
Sbjct: 180 PTLTDIFHSKFDKA 221
>TC14820 homologue to UP|HS80_LYCES (P36181) Heat shock cognate protein 80,
partial (53%)
Length = 1180
Score = 32.3 bits (72), Expect = 0.62
Identities = 16/67 (23%), Positives = 33/67 (48%)
Frame = +1
Query: 1512 ELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNR 1571
E E K I++++ +W + ++++ + + + +E Y A+ YK T W + +
Sbjct: 799 EKEEKKKKKIKEVSHEWSLVNKQKPIWMRKPEEINKEEY--AAFYKSLTNDWEEHLAVKH 972
Query: 1572 EFVSGQL 1578
V GQL
Sbjct: 973 FSVEGQL 993
>TC19742 weakly similar to UP|SDF2_ARATH (Q93ZE8) Stromal cell-derived
factor 2-like protein precursor (SDF2-like protein),
partial (24%)
Length = 474
Score = 32.3 bits (72), Expect = 0.62
Identities = 17/36 (47%), Positives = 21/36 (58%)
Frame = -3
Query: 331 SGRVLHEPEEKIEGDKNEEAEVERDEGVMEERAREP 366
SG E E +IEG++ EE+E E E EER EP
Sbjct: 352 SGGRRREGEFRIEGEEEEESESEESESHEEERCDEP 245
>TC18698
Length = 808
Score = 32.0 bits (71), Expect = 0.81
Identities = 16/42 (38%), Positives = 24/42 (57%)
Frame = -2
Query: 896 FAYKRMPFGLCNAPATFQRCMFAIFSDLIETCIEIFMDDFSV 937
+ Y+ MP GL N T+QR M IF I +E++++D V
Sbjct: 771 YYYQVMPLGLKNI*TTYQRLMDKIFHKQI*KNVEVYVEDMIV 646
>BP070702
Length = 511
Score = 31.6 bits (70), Expect = 1.1
Identities = 15/36 (41%), Positives = 22/36 (60%), Gaps = 1/36 (2%)
Frame = -2
Query: 716 PLEEEKLLRVLRD-HKSALGWTIDDIKGISPAICMH 750
PL +EKL+RVLR +K WT+DD + + + H
Sbjct: 393 PLIDEKLMRVLRRRNKRVYAWTVDDAESMQKMLFEH 286
>TC14821 homologue to UP|AAR12195 (AAR12195) Molecular chaperone Hsp90-1,
partial (29%)
Length = 612
Score = 31.2 bits (69), Expect = 1.4
Identities = 16/67 (23%), Positives = 33/67 (48%)
Frame = +3
Query: 1512 ELEHKAYWAIRKLNFDWKVASEKRLLQLNELDEFRLRAYESASIYKEKTKKWHDRKILNR 1571
E E K I++++ +W + ++++ + + + +E Y A+ YK T W + +
Sbjct: 138 EKEEKKKKKIKEVSHEWSLVNKQKPIWMRKPEEITKDEY--AAFYKSLTNDWEEHLAVKH 311
Query: 1572 EFVSGQL 1578
V GQL
Sbjct: 312 FSVEGQL 332
>TC11985 similar to GB|AAF40452.1|7211981|F13M7 ESTs gb|N65605, gb|N38087,
gb|T20485, gb|T13726, gb|N38339, gb|F15440 and gb|N97201
come from this gene. {Arabidopsis thaliana;}, partial
(88%)
Length = 1334
Score = 30.8 bits (68), Expect = 1.8
Identities = 32/134 (23%), Positives = 55/134 (40%), Gaps = 5/134 (3%)
Frame = +2
Query: 243 KTPTSEQDPL----DQPQ-IVVKTVGKKSLEELIERFIDHTSSNYKSHDMAIKSLESKLG 297
K +DP+ DQ + ++V V KK LEEL + H LES +
Sbjct: 182 KVDAMAEDPIGATTDQTEEMIVPEVDKKLLEELESMGFSTARATRALHYSGNAGLESAVN 361
Query: 298 HLAKQMSDNHQGKFSSETLTLIEQENTAVVSTRSGRVLHEPEEKIEGDKNEEAEVERDEG 357
+ + H+ + + L+ NT VV+++ E + K + + + + +E
Sbjct: 362 WIVE-----HENDSDIDQMPLV-PANTKVVASKPSLTPEELKAKQQELRERARKKKEEEE 523
Query: 358 VMEERAREPTRTRV 371
ER RE R R+
Sbjct: 524 KRMEREREKERIRI 565
>TC17149 similar to GB|AAP37818.1|30725592|BT008459 At1g43860 {Arabidopsis
thaliana;}, partial (48%)
Length = 597
Score = 30.8 bits (68), Expect = 1.8
Identities = 18/50 (36%), Positives = 24/50 (48%)
Frame = +3
Query: 330 RSGRVLHEPEEKIEGDKNEEAEVERDEGVMEERAREPTRTRVVNTHTPEL 379
R+ R + D+ EEA R G E AR+P R RV++ H P L
Sbjct: 12 RNRRGAENVSDASAADRPEEANQRR--GGASEEARQPLRNRVLSQHRPRL 155
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,517,630
Number of Sequences: 28460
Number of extensions: 404765
Number of successful extensions: 1728
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 1713
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1725
length of query: 1649
length of database: 4,897,600
effective HSP length: 103
effective length of query: 1546
effective length of database: 1,966,220
effective search space: 3039776120
effective search space used: 3039776120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0330.2


