

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
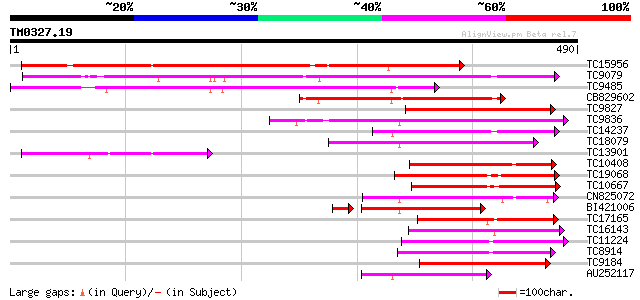
Query= TM0327.19
(490 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 311 2e-85
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 236 5e-63
TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, par... 163 5e-41
CB829602 144 4e-35
TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-... 134 3e-32
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 130 4e-31
TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 127 4e-30
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 125 2e-29
TC13901 similar to UP|Q9FYU7 (Q9FYU7) UDP-glucose:sinapate gluco... 120 5e-28
TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransfe... 114 3e-26
TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 111 2e-25
TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-l... 109 9e-25
CN825072 102 1e-22
BI421006 95 2e-22
TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltran... 100 5e-22
TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltrans... 100 7e-22
TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase... 100 9e-22
TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl tr... 96 1e-20
TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone glu... 94 5e-20
AU252117 92 1e-19
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 311 bits (796), Expect = 2e-85
Identities = 163/387 (42%), Positives = 238/387 (61%), Gaps = 4/387 (1%)
Frame = +2
Query: 11 LHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTN 70
LH++LV+F AQGHINPLLRL K L A+G VTLATT+ KS T TD PT
Sbjct: 35 LHILLVSFPAQGHINPLLRLAKCLAAKGSSVTLATTQTAG----KSMRTATNITDKAPTP 202
Query: 71 FTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLACII 130
+ + FF DGL P+ D + Y+ +E +G + +IK H +N ++++CII
Sbjct: 203 ISDGSLKFEFFDDGLGPDDDSIRGNLSLYLQQLELVGRQQILKMIKEHADSN-RQISCII 379
Query: 131 NNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPTLENPELNVQLPGLPL 190
NNPF+PWV DVA E IP A LWIQ A++ YY +++N FP+ E+P +VQLP +
Sbjct: 380 NNPFLPWVVDVAEEQGIPSALLWIQSSAVFTAYYSYFHNLVTFPSKEDPLADVQLPSTSI 559
Query: 191 -LKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKEVIDSMAETYPVI 249
LK ++P F+ P+ + L ++ + K++ K +L +++ ELE + ID +++T+ +
Sbjct: 560 VLKHCEIPDFLHPSCTYPFLGTLILEQFKNLNKTFCILVDTYEELEHDFIDYLSKTFLIR 739
Query: 250 PVGPLLPLSLLGVDENEDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLES 309
PVGPL S ++ + + K D C+EWLN +P SSV+Y+SFGS++ L ++++
Sbjct: 740 PVGPLFKSSQA---KSAAIRGDFMK-SDDCIEWLNSKPQSSVVYVSFGSIVYLPQEQVDE 907
Query: 310 IATALKNSNCKFLWVIK---KQDGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIA 366
IA LK S FLWV+K + G LP GF EET RG VV W PQ +VL HP+++
Sbjct: 908 IAFGLKKSQVSFLWVMKPPPEVTGLQPHVLPYGFLEETGERGRVVQWSPQEEVLAHPSVS 1087
Query: 367 CFLTHCGWNSMLEAIAAGKPMIAYPQW 393
CFLTHCGWNS +EA+ +G P++ +P W
Sbjct: 1088CFLTHCGWNSSMEALTSGVPVLTFPAW 1168
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 236 bits (603), Expect = 5e-63
Identities = 148/482 (30%), Positives = 244/482 (49%), Gaps = 18/482 (3%)
Frame = +1
Query: 12 HVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATTELVYHRVFKSSSTTAATTDTIPTNF 71
H +L QGHINPL +L K L RG +T TE + R+ KS A D +P +F
Sbjct: 19 HAVLTPSPLQGHINPLFQLAKLLHLRGFHITFVHTEYNHKRLIKSRGPNAL--DGLP-DF 189
Query: 72 TTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSKKLA---- 127
I DGL+ + + N + L+ + S L
Sbjct: 190 VFETI-----PDGLEDGDGEVSQDLYSLCDSIRNKCHLPFQDLLAKLHRSASAGLVPPVT 354
Query: 128 CIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQ--FPT-----LENPE 180
C++++ + + A + ++P L + + + HF ++ P L N
Sbjct: 355 CLVSDLLMTFTIQAAQQLQLPILLLCPASASTFMSFLHFQTLLDKGVIPLKDESYLTNGY 534
Query: 181 LNVQ---LPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLKWVLANSFYELEKE 237
L+ + +PG+ + +DLP FI T+P + + + ++ K+ ++ N+F ELE++
Sbjct: 535 LDTKVDWIPGMQNFRLKDLPDFIRTTDPNDIMLEFMVEVADRAKRASAIVFNTFNELERD 714
Query: 238 VIDSMAETYPVI-PVGPLLPLSLLGVDENE--DVGIEMWKPQDSCLEWLNDQPPSSVIYI 294
V+ +++ P + P+GP P L +N+ +G +WK CL+WL + P SV+Y+
Sbjct: 715 VLSALSTMLPSLYPIGPF-PSFLNQAPQNQLASLGSNLWKEDTKCLQWLESKEPGSVVYV 891
Query: 295 SFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKD-SLPLPQGFKEETKNRGMVVPW 353
+FGS+ V+S ++L A L N FLW+I+ D S L F +ET NRG++ W
Sbjct: 892 NFGSITVMSPEQLLEFAWGLANGKKPFLWIIRPDLVADGSAILSPKFVDETSNRGLIASW 1071
Query: 354 CPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRL 413
CPQ KVL HP++ FLTHCGWNS +E+I AG PM+ +P DQPTN + + + + +GI +
Sbjct: 1072CPQEKVLNHPSVGGFLTHCGWNSTIESICAGVPMLCWPFVADQPTNCRYICNEWDIGIEV 1251
Query: 414 EQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDE 473
+ + V+ E+E I E++ G K +++++ ELK+ A E GG S N+ + E
Sbjct: 1252DTN----VKREEVENLIHELMVGDKGKKMRQRTMELKKKAEEDTRPGGCSYMNLDKLIKE 1419
Query: 474 IL 475
+L
Sbjct: 1420VL 1425
>TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, partial (75%)
Length = 1195
Score = 163 bits (413), Expect = 5e-41
Identities = 118/391 (30%), Positives = 193/391 (49%), Gaps = 20/391 (5%)
Frame = +1
Query: 1 MTSSESRAEELHVMLVAFSAQGHINPLLRLGKSLLARG-LQVTLATTELVYHRVFKSSST 59
M S + A++ HV+ + + AQGHINP+L+L K L +G VT TE + R+ KS
Sbjct: 58 MGSLGTAAKKPHVVCIPYPAQGHINPMLKLAKLLHFKGGFHVTFVNTEYNHKRLLKSRGP 237
Query: 60 TAATTDTIPTNFTTNGIHVLFFS---DGL-DPNPDQTQLTPDDYMALVENLGPINLSTLI 115
+ NG+ F DGL + + D TQ P ++ + P L
Sbjct: 238 DSL-----------NGLPSFRFETIPDGLPETDVDVTQDIPSLCISTRKTCLPHFKKLLS 384
Query: 116 KTHFINNS-KKLACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQ-- 172
K + +++ + CI+++ + + D A E IP W + Y + +
Sbjct: 385 KLNDVSSDVPPVTCIVSDGCMSFTLDAAIELNIPEVLFWTTSACGFMGYVQYRELIEKGI 564
Query: 173 FPTLENPELN--------VQLPGLPLLKPQDLPSFILPTNPFGALSKVLADMLKDMKKLK 224
P ++ ++ LPG+ ++ +DLPSF+ T+P + L + K
Sbjct: 565 IPLKDSSDITNGYLETTIEWLPGMKNIRLKDLPSFLRTTDPNDKMLDFLTGECQRALKAS 744
Query: 225 WVLANSFYELEKEVIDSMAETYP-VIPVGPLLPLSLLGVDEN-EDVGIEMWKPQDSCLEW 282
++ N+F LE +V+++ + P V +GPL L D+N +G +WK CL+W
Sbjct: 745 AIILNTFDALEHDVLEAFSSILPPVYSIGPLHLLIKDVTDKNLNSLGSNLWKEDSECLKW 924
Query: 283 LNDQPPSSVIYISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQ--DGKDSLPLPQGF 340
L+ + P+SV+Y++FGS+ V++++++ A L NSN FLWVI+ GK ++ LP+ F
Sbjct: 925 LDTKEPNSVVYVNFGSITVMTSEQMVEFAWGLANSNKTFLWVIRPDLVAGKHAV-LPEEF 1101
Query: 341 KEETKNRGMVVPWCPQTKVLVHPAIACFLTH 371
T +RG + W PQ VL HPAI FLTH
Sbjct: 1102VAATNDRGRLSSWTPQEDVLTHPAIGGFLTH 1194
>CB829602
Length = 542
Score = 144 bits (362), Expect = 4e-35
Identities = 74/182 (40%), Positives = 117/182 (63%), Gaps = 4/182 (2%)
Frame = +1
Query: 251 VGPLLPLSLLGVDEN--EDVGIEMWKPQDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLE 308
+GPL L L V EN E + +WK + CL+WL+ Q PSSV+Y++FGS+I ++ ++L
Sbjct: 7 IGPL-DLLLNQVTENRIESIKCNLWKEEPECLKWLDSQEPSSVLYVNFGSVINMTPQQLV 183
Query: 309 SIATALKNSNCKFLWVIKKQ--DGKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIA 366
+A + NS KF+WVI+ +G+ S+ LP+ ETK+RG+++ WCPQ +VL H A+
Sbjct: 184 ELAWGIANSKKKFIWVIRPDLVEGEASIVLPE-IVAETKDRGIMLSWCPQEQVLKHSALG 360
Query: 367 CFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVETGEL 426
FLTHCGWNS +E+I++G P+I P + DQ N++ + + G+ + D V+ E+
Sbjct: 361 GFLTHCGWNSTIESISSGVPLICSPFFNDQFVNSRYICSEWDFGMEMNSDD---VKRDEV 531
Query: 427 ER 428
E+
Sbjct: 532 EK 537
>TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (42%)
Length = 611
Score = 134 bits (337), Expect = 3e-32
Identities = 60/129 (46%), Positives = 84/129 (64%)
Frame = +2
Query: 343 ETKNRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKL 402
ET +G++V WCPQ VL H A+ CFLTHCGWNS LE+++ G P+IA P WTD TN KL
Sbjct: 11 ETTEKGLIVTWCPQLLVLKHQALGCFLTHCGWNSTLESVSLGVPVIAMPLWTDPVTNGKL 190
Query: 403 VSDVFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGS 462
+ DV++ G+R D V ++ I+EI+ K E+K NA + A++++A+GG
Sbjct: 191 IVDVWKTGVRAVADEKEIVRRETIKNCIKEIIETEKINEIKSNAIKWMNLAKDSLAEGGR 370
Query: 463 SDRNIQTFV 471
SD+NI FV
Sbjct: 371 SDKNIAEFV 397
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 130 bits (328), Expect = 4e-31
Identities = 82/275 (29%), Positives = 142/275 (50%), Gaps = 16/275 (5%)
Frame = +1
Query: 225 WVLANSFYELEKEVIDSMAETY-------PVIPVGPLLPLSLLGVDENEDVGIEMWKPQD 277
WV ++ F DSM Y P +GP+L LS + GI +
Sbjct: 211 WVNSDGFLFNTVADFDSMGLRYVARKLNRPAWAIGPVL-LSTGSGSRGKGGGIS----PE 375
Query: 278 SCLEWLNDQPPSSVIYISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLP-- 335
C +WL+ +P +SV+++SFGS+ +SA ++ +ATAL S F+WV++ G D
Sbjct: 376 LCRKWLDTKPSNSVLFVSFGSMNTISASQMMQLATALDRSGRNFIWVVRPPIGFDINSEF 555
Query: 336 -----LPQGFKEETKNRGMVV-PWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIA 389
LP+GF +G+VV W PQ ++L H A++ FL+HCGWNS+LE+++ G P++
Sbjct: 556 RAEEWLPEGFLGRVAKKGLVVHDWAPQVEILSHGAVSAFLSHCGWNSVLESLSHGVPILG 735
Query: 390 YPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVETGELERAIEEIVSGPKS-EELKRNAAE 448
+P +Q N K++ + V + + + V +L IE +++ +S ++++NA
Sbjct: 736 WPMAAEQFFNCKMLEEELGVCVEVARGKRCEVRHEDLVEKIELVMNEAESGVKIRKNAGN 915
Query: 449 LKRAAREAVADGGSSDRNIQTFVDEILATRLDVME 483
++ R+AV D + +DE L+ + + E
Sbjct: 916 IREMIRDAVRDEDGYKGSSVRAIDEFLSAAMSLNE 1020
>TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (27%)
Length = 999
Score = 127 bits (319), Expect = 4e-30
Identities = 61/164 (37%), Positives = 97/164 (58%), Gaps = 2/164 (1%)
Frame = +2
Query: 314 LKNSNCKFLWVIKKQD--GKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIACFLTH 371
+ NS FLW+++ G+DS+ +PQ F +E K RG + WC Q +VL HP+I FLTH
Sbjct: 197 IANSKVPFLWILRPDVVMGEDSINVPQEFLDEIKGRGYIASWCFQEEVLSHPSIGAFLTH 376
Query: 372 CGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVETGELERAIE 431
CGWNS +E I+AG P+I +P +++Q TN++ + +G+ + D V+ E+ +
Sbjct: 377 CGWNSTIEGISAGLPLICWPFFSEQHTNSRYACTTWEIGMEVNHD----VKRDEITTLVN 544
Query: 432 EIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNIQTFVDEIL 475
E++ G K +E+++ E K+ A EA GGSS + + E L
Sbjct: 545 EMMKGEKGKEMRKKGLEWKKKAIEATGLGGSSYNDFHKLIKESL 676
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 125 bits (314), Expect = 2e-29
Identities = 67/196 (34%), Positives = 108/196 (54%), Gaps = 14/196 (7%)
Frame = +3
Query: 276 QDSCLEWLNDQPPSSVIYISFGSLIVLSAKKLESIATALKNSNCKFLWVIKKQDGKDSLP 335
QD L WL+ QP SVIY+SFGS + ++ IA L+ S +F+WV++ + D+
Sbjct: 51 QDEILRWLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGLELSQHRFVWVVRPPNEADASA 230
Query: 336 -------------LPQGFKEETKNRGMVVP-WCPQTKVLVHPAIACFLTHCGWNSMLEAI 381
LP+GF T+ G+VVP W PQ ++L HPA F+T CGWNS+LE++
Sbjct: 231 TFFGFGNEMALNYLPEGFMNRTREFGVVVPMWAPQAEILGHPATGGFVTPCGWNSVLESV 410
Query: 382 AAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEE 441
G PM+A+P +++Q NA ++S+ V +R++ G V ++ + ++ +
Sbjct: 411 LNGVPMVAWPLYSEQKMNAYMLSEELGVAVRVKVAEGGVVCRDQVVDMVRRVMVSEEGMG 590
Query: 442 LKRNAAELKRAAREAV 457
+K ELK + EA+
Sbjct: 591 MKDRVRELKLSGDEAL 638
>TC13901 similar to UP|Q9FYU7 (Q9FYU7) UDP-glucose:sinapate
glucosyltransferase , partial (16%)
Length = 558
Score = 120 bits (301), Expect = 5e-28
Identities = 72/171 (42%), Positives = 94/171 (54%), Gaps = 6/171 (3%)
Frame = +3
Query: 11 LHVMLVAFSAQGHINPLLRLGKSLLARGLQVTLATT-ELVYHRVFKSSSTTAATTDTI-- 67
+HV+LV+F AQGHI+PLLRLGK L A+GL VT +TT + V +++TT AT +
Sbjct: 45 IHVLLVSFPAQGHISPLLRLGKHLAAKGLFVTFSTTRQKGEEMVVAAAATTTATANGTSS 224
Query: 68 ---PTNFTTNGIHVLFFSDGLDPNPDQTQLTPDDYMALVENLGPINLSTLIKTHFINNSK 124
PT + + DGL P T+ DY G L LIK H N +
Sbjct: 225 PPPPTPIGDGFLQFDLYDDGL-PEDHPTRNDLKDYSLQXAEYGKKILPQLIKKHADAN-R 398
Query: 125 KLACIINNPFVPWVADVAAEFKIPCACLWIQPCALYAIYYHFYNNTNQFPT 175
++CIINNPFV WV +VAAE IPCA WI CA + +H++ FPT
Sbjct: 399 PVSCIINNPFVSWVCEVAAELDIPCAMYWIHACATFVACHHYFQKLTPFPT 551
>TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (19%)
Length = 571
Score = 114 bits (286), Expect = 3e-26
Identities = 49/127 (38%), Positives = 82/127 (63%)
Frame = +1
Query: 346 NRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSD 405
++G VV W PQ K+L HPAIACF++HCGWNS +E + A P + +P TDQ N + D
Sbjct: 10 SKGKVVKWGPQKKILNHPAIACFISHCGWNSTIEGVHASVPFLCWPFSTDQFLNKSFICD 189
Query: 406 VFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDR 465
V+++G+ L++D +G + GE+ +E+++ +E+LK + +LK +A+GG S
Sbjct: 190 VWKIGLGLDKDENGIIPKGEIRTKVEQVI---VNEDLKARSLKLKEITINNLAEGGQSSN 360
Query: 466 NIQTFVD 472
N++ F++
Sbjct: 361 NLKKFIN 381
>TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (26%)
Length = 565
Score = 111 bits (278), Expect = 2e-25
Identities = 55/143 (38%), Positives = 89/143 (61%)
Frame = +1
Query: 333 SLPLPQGFKEETKNRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQ 392
S+ L F E +RG++ WC Q VL HP+I FLTHCGWNS +E+I+AG PM+ +P
Sbjct: 7 SVILSSEFINEISSRGLIASWCSQEHVLNHPSIGGFLTHCGWNSTIESISAGVPMLCWPF 186
Query: 393 WTDQPTNAKLVSDVFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRA 452
+ DQ TN + + + + +G+++ D++G E E+E+ I E++ G K +++++ ELK+
Sbjct: 187 FADQLTNCRSICNEWDIGMQI--DTNGKRE--EVEKLINELMVGEKGKKMRQKTMELKKR 354
Query: 453 AREAVADGGSSDRNIQTFVDEIL 475
A E GG S N+ + E+L
Sbjct: 355 AEEDTRPGGCSYMNLDQVIKEVL 423
>TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-like
protein, partial (20%)
Length = 824
Score = 109 bits (273), Expect = 9e-25
Identities = 58/129 (44%), Positives = 81/129 (61%)
Frame = +1
Query: 348 GMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVF 407
G V W PQ +VL HPAI F THCGWNS LE++ G PMI P + DQ NAK VSDV+
Sbjct: 7 GTSVKWAPQEQVLKHPAIGAFWTHCGWNSTLESVCEGVPMICSPCFGDQKVNAKYVSDVW 186
Query: 408 RVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNI 467
+VG++L Q+ G GE+E+ I ++ G ++ E++ N +LK A +++GGSS +
Sbjct: 187 KVGVQL-QNKPG---RGEIEKTISLLMLGDEAYEIRGNILKLKEKANVCLSEGGSSYCFL 354
Query: 468 QTFVDEILA 476
V EIL+
Sbjct: 355 DRLVSEILS 381
>CN825072
Length = 712
Score = 102 bits (254), Expect = 1e-22
Identities = 58/194 (29%), Positives = 100/194 (50%), Gaps = 25/194 (12%)
Frame = -3
Query: 306 KLESIATALKNSNCKFLWVIKKQDGKDSLP-------------LPQGFKEETKNRGMVVP 352
++ IA A++NS +F+W ++K K S+ LP+GF + T G V+
Sbjct: 683 QITEIARAVENSGVRFVWSLRKPXLKGSMVGPSDYSVDDLASVLPEGFLDRTTGIGRVIG 504
Query: 353 WCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIR 412
W PQT+VL HPA F++HCGWNS LE+I G P+ +P + +Q TNA ++ ++ +
Sbjct: 503 WAPQTRVLTHPATGGFVSHCGWNSTLESIYFGVPIATWPLYAEQQTNAFVLVRELKIAVE 324
Query: 413 LEQDSDGFVETG--------ELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSS- 463
+ D + G ++E I ++ K E+++ E+ +R+ + +GG S
Sbjct: 323 ISLDYRVEINGGPNYLLTADKIEGGIRSVLD--KDGEVRKRVKEMSEKSRKTLLEGGCSY 150
Query: 464 ---DRNIQTFVDEI 474
DR I F+ ++
Sbjct: 149 SYLDRLIDYFMHQV 108
>BI421006
Length = 519
Score = 95.1 bits (235), Expect(2) = 2e-22
Identities = 43/116 (37%), Positives = 72/116 (62%), Gaps = 9/116 (7%)
Frame = +2
Query: 305 KKLESIATALKNSNCKFLWVIKKQDGKDSLP--------LPQGFKEETKNRGMVV-PWCP 355
+++E IA L+ S KF+WV++ D D LP+GF++ + G+VV W P
Sbjct: 128 EQIEQIANGLEQSKQKFIWVLRDADKGDIFDGDKVKERGLPKGFEKRVEGMGLVVRDWAP 307
Query: 356 QTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGI 411
Q ++L HP+ F++HCGWNS +E+++ G P+ A+P +DQP N L+++V +V +
Sbjct: 308 QLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPRNTVLITEVLKVAL 475
Score = 27.7 bits (60), Expect(2) = 2e-22
Identities = 10/18 (55%), Positives = 14/18 (77%)
Frame = +3
Query: 280 LEWLNDQPPSSVIYISFG 297
+EWL+ Q SV+Y+SFG
Sbjct: 54 MEWLDRQEQKSVLYVSFG 107
>TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltransferase,
partial (24%)
Length = 546
Score = 100 bits (249), Expect = 5e-22
Identities = 48/130 (36%), Positives = 80/130 (60%), Gaps = 8/130 (6%)
Frame = +1
Query: 353 WCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGI- 411
W PQ +L H AI F+THCGWNS LE +AAG PM+ +P +Q N KLV++V ++G+
Sbjct: 13 WAPQVLILEHEAIGAFVTHCGWNSTLEGVAAGVPMVTWPIAAEQFYNEKLVTEVLKIGVP 192
Query: 412 -------RLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSD 464
R+ + DG V++ +E+ ++ I+ G ++EE++ L + A+ AV +GGSS
Sbjct: 193 VGAKKWGRMVLEGDG-VKSDAVEKGVKRIMEGEEAEEMRNRVKVLSQQAQWAVEEGGSSY 369
Query: 465 RNIQTFVDEI 474
++ ++E+
Sbjct: 370 SDLNALIEEL 399
>TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (30%)
Length = 604
Score = 100 bits (248), Expect = 7e-22
Identities = 49/146 (33%), Positives = 86/146 (58%), Gaps = 11/146 (7%)
Frame = +1
Query: 345 KNRGMVVP-WCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLV 403
+ +GM++ W PQ +L H A+ F+THCGWNS +EA++AG PMI +P +Q N KL+
Sbjct: 1 REKGMIIRGWVPQVVILGHRAVGAFVTHCGWNSTVEAVSAGVPMITWPVVGEQFYNEKLI 180
Query: 404 SDVFRVGIRLEQDS---------DGFVETGELERAIEEIV-SGPKSEELKRNAAELKRAA 453
++V +G+ + + + V +E+A+ ++ G + E+++R A E A
Sbjct: 181 TEVRGIGVEVGAEECCLMGFDKREKLVSRESIEKAVRRLMDGGDEGEQIRRRAQEYGEKA 360
Query: 454 REAVADGGSSDRNIQTFVDEILATRL 479
R+AV +GGSS +N+ +D++ R+
Sbjct: 361 RQAVEEGGSSHKNLTALIDDLKRLRV 438
>TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (19%)
Length = 596
Score = 99.8 bits (247), Expect = 9e-22
Identities = 52/145 (35%), Positives = 85/145 (57%)
Frame = +3
Query: 339 GFKEETKNRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPT 398
GF E TK++G VV W PQ ++L H ++ FLTH GWNS+LE I G PMI P + DQ
Sbjct: 6 GFLERTKSQGKVVSWAPQMEILKHASVGVFLTHGGWNSVLECIVGGVPMIGRPFFGDQRI 185
Query: 399 NAKLVSDVFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVA 458
N +++ + VG+ L+ +G + + + ++ ++G + + +++ AELK A +AV
Sbjct: 186 NIWMLAKLLGVGVGLQ---NGVLAKETILKTLKSTMTGEEGKVMRQKMAELKEMAWKAVE 356
Query: 459 DGGSSDRNIQTFVDEILATRLDVME 483
GSS +N+ T + IL L ++
Sbjct: 357 PDGSSTKNLCTLMQIILG*NLRTLD 431
>TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl transferase,
partial (32%)
Length = 627
Score = 95.9 bits (237), Expect = 1e-20
Identities = 44/136 (32%), Positives = 79/136 (57%)
Frame = +2
Query: 336 LPQGFKEETKNRGMVVPWCPQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTD 395
LP GF E TKN G +VPW PQT++L H ++ F+THCG NS+ E+I+ G PMI P + D
Sbjct: 29 LPNGFIERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVPMICRPFFGD 208
Query: 396 QPTNAKLVSDVFRVGIRLEQDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAARE 455
+++ D++ +G+++E G L +++ I+ + + ++ A ++K+ +
Sbjct: 209 HGMAGRIIEDIWEIGVKIE---GGVFTQDGLVKSLNLILVQEEGKIMREKAQKVKKTVLD 379
Query: 456 AVADGGSSDRNIQTFV 471
A G + ++ +T V
Sbjct: 380 AAGPQGKATQDFKTLV 427
>TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (26%)
Length = 643
Score = 94.0 bits (232), Expect = 5e-20
Identities = 44/113 (38%), Positives = 71/113 (61%)
Frame = +3
Query: 355 PQTKVLVHPAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLE 414
PQ KVL H +I FL+HCGWNS+LE++A G P++A+P + +Q NA LV++ +V +R +
Sbjct: 6 PQAKVLSHASIGGFLSHCGWNSVLESVANGVPLVAWPLYAEQRMNAVLVTEDVKVALRPK 185
Query: 415 QDSDGFVETGELERAIEEIVSGPKSEELKRNAAELKRAAREAVADGGSSDRNI 467
+G VE E+ R + ++ G + ++L+ +LK AA + + GSS I
Sbjct: 186 VGENGLVELVEIARVVRTLMQGEEGKKLRCKMKDLKEAAATTLKENGSSTNQI 344
>AU252117
Length = 350
Score = 92.4 bits (228), Expect = 1e-19
Identities = 41/114 (35%), Positives = 64/114 (55%), Gaps = 2/114 (1%)
Frame = +3
Query: 305 KKLESIATALKNSNCKFLWVIKKQD--GKDSLPLPQGFKEETKNRGMVVPWCPQTKVLVH 362
K + + NS FLW+++ G + + LPQ F +E K+RG + WC Q +VL H
Sbjct: 6 KHFKEFVWGIANSKVPFLWIMRPDVVMGGEFINLPQEFLDEIKDRGYIASWCNQEQVLAH 185
Query: 363 PAIACFLTHCGWNSMLEAIAAGKPMIAYPQWTDQPTNAKLVSDVFRVGIRLEQD 416
P+I FLTHC WNS + +++G PMI +P + +Q TN + + G+ + D
Sbjct: 186 PSIGVFLTHCDWNSTVVCVSSGVPMICWPFFAEQQTNCRYACTTWGTGMDINHD 347
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,895,006
Number of Sequences: 28460
Number of extensions: 135138
Number of successful extensions: 868
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 833
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 838
length of query: 490
length of database: 4,897,600
effective HSP length: 94
effective length of query: 396
effective length of database: 2,222,360
effective search space: 880054560
effective search space used: 880054560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0327.19


