

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0322.4
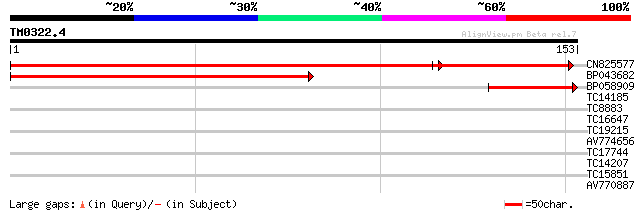
(153 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CN825577 222 2e-75
BP043682 169 3e-43
BP058909 60 2e-10
TC14185 similar to UP|Q7Y256 (Q7Y256) Beta-cyanoalanine synthase... 33 0.028
TC8883 30 0.14
TC16647 weakly similar to UP|PAP4_CARPA (P05994) Papaya proteina... 28 0.54
TC19215 weakly similar to UP|Q9XIN9 (Q9XIN9) At2g27320 protein, ... 26 3.5
AV774656 26 3.5
TC17744 25 4.5
TC14207 similar to UP|RK15_PEA (P31165) 50S ribosomal protein L1... 25 4.5
TC15851 25 5.9
AV770887 25 7.7
>CN825577
Length = 706
Score = 222 bits (565), Expect(2) = 2e-75
Identities = 108/117 (92%), Positives = 109/117 (92%)
Frame = +1
Query: 1 LIL*PIMEIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPW 60
LIL*PIMEIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTY EVIDEIYNQVDHVEPW
Sbjct: 136 LIL*PIMEIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYHEVIDEIYNQVDHVEPW 315
Query: 61 MAGNCRGPSTFFCLLY*FFTMMLTVKQMHGLLKHPDSPYIRVVGFLYLRYVPDPNTV 117
M GNCRGPST FCLLY FFTM LTVKQMHGLLKHPDSPYIR VGFLYLRYV DP T+
Sbjct: 316 MTGNCRGPSTSFCLLYKFFTMKLTVKQMHGLLKHPDSPYIRAVGFLYLRYVADPKTL 486
Score = 75.5 bits (184), Expect(2) = 2e-75
Identities = 35/38 (92%), Positives = 37/38 (97%)
Frame = +3
Query: 115 NTV*LV*TIYKG**GIFSWI*WTNDYNGCIYP*FAPWT 152
++V LV*TIYKG**GIFSWI*WTNDYNGCIYP*FAPWT
Sbjct: 480 DSVELV*TIYKG**GIFSWI*WTNDYNGCIYP*FAPWT 593
>BP043682
Length = 499
Score = 169 bits (427), Expect = 3e-43
Identities = 82/82 (100%), Positives = 82/82 (100%)
Frame = +2
Query: 1 LIL*PIMEIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPW 60
LIL*PIMEIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPW
Sbjct: 254 LIL*PIMEIQTSGRPIESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQVDHVEPW 433
Query: 61 MAGNCRGPSTFFCLLY*FFTMM 82
MAGNCRGPSTFFCLLY*FFTMM
Sbjct: 434 MAGNCRGPSTFFCLLY*FFTMM 499
>BP058909
Length = 381
Score = 60.1 bits (144), Expect = 2e-10
Identities = 24/24 (100%), Positives = 24/24 (100%)
Frame = -2
Query: 130 IFSWI*WTNDYNGCIYP*FAPWTG 153
IFSWI*WTNDYNGCIYP*FAPWTG
Sbjct: 146 IFSWI*WTNDYNGCIYP*FAPWTG 75
>TC14185 similar to UP|Q7Y256 (Q7Y256) Beta-cyanoalanine synthase, partial
(96%)
Length = 1506
Score = 32.7 bits (73), Expect = 0.028
Identities = 15/27 (55%), Positives = 17/27 (62%)
Frame = -2
Query: 91 LLKHPDSPYIRVVGFLYLRYVPDPNTV 117
LL +PYI +GFL LRY P P TV
Sbjct: 857 LLSDGSTPYILTLGFLDLRYCPTPETV 777
>TC8883
Length = 602
Score = 30.4 bits (67), Expect = 0.14
Identities = 12/25 (48%), Positives = 19/25 (76%)
Frame = +1
Query: 68 PSTFFCLLY*FFTMMLTVKQMHGLL 92
P +F CL+Y FF + ++V ++HGLL
Sbjct: 460 PISFCCLMYFFFKVDVSVGELHGLL 534
>TC16647 weakly similar to UP|PAP4_CARPA (P05994) Papaya proteinase IV
precursor (PPIV) (Papaya peptidase B) (Glycyl
endopeptidase) , partial (8%)
Length = 616
Score = 28.5 bits (62), Expect = 0.54
Identities = 17/61 (27%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Frame = -2
Query: 5 PIMEIQTSGRPIESLLEKVLCMNILSSDYFKEL--YRLKTYLEVIDEIYNQVDHVEPWMA 62
P P+E L+ + +C I + FKEL + L+ + + +I + D VEP
Sbjct: 420 PFKSFLEDTEPVEFLIFRRVCFVIFLAPAFKELSGFPLRVEMAIGIQIETETDRVEPQNV 241
Query: 63 G 63
G
Sbjct: 240 G 238
>TC19215 weakly similar to UP|Q9XIN9 (Q9XIN9) At2g27320 protein, partial
(12%)
Length = 548
Score = 25.8 bits (55), Expect = 3.5
Identities = 13/34 (38%), Positives = 19/34 (55%), Gaps = 3/34 (8%)
Frame = +1
Query: 35 KELYRLKTY---LEVIDEIYNQVDHVEPWMAGNC 65
+ELY+ + LE+ID YN+V +M NC
Sbjct: 247 RELYKNSAFMSNLEIIDSNYNEVRVNPKYMKSNC 348
>AV774656
Length = 456
Score = 25.8 bits (55), Expect = 3.5
Identities = 10/23 (43%), Positives = 15/23 (64%)
Frame = -1
Query: 90 GLLKHPDSPYIRVVGFLYLRYVP 112
G ++H P+IR++GF Y R P
Sbjct: 333 GPVRHTPYPFIRIIGFDYQRPSP 265
>TC17744
Length = 830
Score = 25.4 bits (54), Expect = 4.5
Identities = 13/33 (39%), Positives = 20/33 (60%), Gaps = 4/33 (12%)
Frame = -2
Query: 35 KELY-RLKTYLEVIDEI---YNQVDHVEPWMAG 63
+ELY RL Y ++ D++ Y+Q +H PW G
Sbjct: 340 RELYPRLLAYNQIEDQMVYFYHQHNHPHPWQEG 242
>TC14207 similar to UP|RK15_PEA (P31165) 50S ribosomal protein L15,
chloroplast precursor (CL15) (Fragment), partial (89%)
Length = 1201
Score = 25.4 bits (54), Expect = 4.5
Identities = 11/37 (29%), Positives = 19/37 (50%), Gaps = 3/37 (8%)
Frame = +2
Query: 42 TYLEVIDEIYNQVDHV---EPWMAGNCRGPSTFFCLL 75
T+ I +I+ ++ + + W A CR PS CL+
Sbjct: 1070 TFCSAISKIFREIQSLIKCKFWFANICRKPSVVVCLV 1180
>TC15851
Length = 684
Score = 25.0 bits (53), Expect = 5.9
Identities = 18/39 (46%), Positives = 21/39 (53%)
Frame = +1
Query: 16 IESLLEKVLCMNILSSDYFKELYRLKTYLEVIDEIYNQV 54
IE + VL +N SS K LYRLKT L D Y+ V
Sbjct: 295 IEEPGKVVLTINNTSSKKKKLLYRLKTKLTPSD*EYHHV 411
>AV770887
Length = 510
Score = 24.6 bits (52), Expect = 7.7
Identities = 10/23 (43%), Positives = 14/23 (60%)
Frame = +3
Query: 39 RLKTYLEVIDEIYNQVDHVEPWM 61
R KT + I ++Q DHV PW+
Sbjct: 39 RSKTIIFYIQGTWSQRDHVTPWI 107
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.352 0.162 0.599
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,895,384
Number of Sequences: 28460
Number of extensions: 39778
Number of successful extensions: 305
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 305
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 305
length of query: 153
length of database: 4,897,600
effective HSP length: 82
effective length of query: 71
effective length of database: 2,563,880
effective search space: 182035480
effective search space used: 182035480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0322.4


