

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0322.12
(601 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
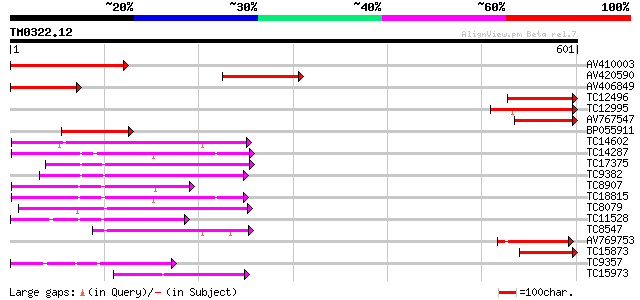
Score E
Sequences producing significant alignments: (bits) Value
AV410003 192 2e-49
AV420590 179 1e-45
AV406849 147 5e-36
TC12496 similar to UP|Q8SA64 (Q8SA64) NIMA-related protein kinas... 147 6e-36
TC12995 136 1e-32
AV767547 129 1e-30
BP055911 122 2e-28
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 118 3e-27
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 115 3e-26
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 114 3e-26
TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein ... 105 3e-23
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 100 5e-22
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 96 2e-20
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 94 6e-20
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 94 8e-20
TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated pro... 88 3e-18
AV769753 86 2e-17
TC15873 similar to UP|Q9LXP3 (Q9LXP3) Protein kinase-like protei... 82 2e-16
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 82 2e-16
TC15973 homologue to GB|AAL15278.1|16323087|AY057647 AT3g01490/F... 80 9e-16
>AV410003
Length = 428
Score = 192 bits (487), Expect = 2e-49
Identities = 82/126 (65%), Positives = 111/126 (88%)
Frame = +3
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
M+QYE++EQIG+G+FG+A+LV HK E+K YVLKKIRLARQ++R RRSAHQEM LI+++++
Sbjct: 51 MDQYEIMEQIGRGAFGAAILVNHKAERKKYVLKKIRLARQTERCRRSAHQEMALIARIQH 230
Query: 61 PFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYL 120
P+IVE+K++WVEKGC+VCI+ GYCE GD+A+ +KK NG PEEKLCKW QLL+A++YL
Sbjct: 231 PYIVEFKEAWVEKGCYVCIVTGYCEGGDMAELMKKQNGAYIPEEKLCKWFTQLLLAVEYL 410
Query: 121 HVNHIL 126
H N++L
Sbjct: 411 HSNYVL 428
>AV420590
Length = 260
Score = 179 bits (453), Expect = 1e-45
Identities = 86/86 (100%), Positives = 86/86 (100%)
Frame = +3
Query: 226 MYSSAFRGLVKSMLRKNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESN 285
MYSSAFRGLVKSMLRKNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESN
Sbjct: 3 MYSSAFRGLVKSMLRKNPELRPPASELLNHPHLQPYILKIHLKLNSPRRSTFPFQWPESN 182
Query: 286 YMRRTRFVVPDSVSTLSDQDKCLSLS 311
YMRRTRFVVPDSVSTLSDQDKCLSLS
Sbjct: 183 YMRRTRFVVPDSVSTLSDQDKCLSLS 260
>AV406849
Length = 259
Score = 147 bits (371), Expect = 5e-36
Identities = 72/76 (94%), Positives = 73/76 (95%)
Frame = +1
Query: 1 MEQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRN 60
MEQYEVLEQIGKGSF SALLVRHKHE K YVLKKIRLARQ+DRTRRSAHQEMELISKVRN
Sbjct: 31 MEQYEVLEQIGKGSFASALLVRHKHENKKYVLKKIRLARQTDRTRRSAHQEMELISKVRN 210
Query: 61 PFIVEYKDSWVEKGCF 76
PFIVEYKDSWVEKGCF
Sbjct: 211 PFIVEYKDSWVEKGCF 258
>TC12496 similar to UP|Q8SA64 (Q8SA64) NIMA-related protein kinase, partial
(10%)
Length = 770
Score = 147 bits (370), Expect = 6e-36
Identities = 74/74 (100%), Positives = 74/74 (100%)
Frame = +3
Query: 528 SIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRE 587
SIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRE
Sbjct: 3 SIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRE 182
Query: 588 TAIWLAKSFKETVI 601
TAIWLAKSFKETVI
Sbjct: 183 TAIWLAKSFKETVI 224
>TC12995
Length = 513
Score = 136 bits (342), Expect = 1e-32
Identities = 72/95 (75%), Positives = 77/95 (80%), Gaps = 3/95 (3%)
Frame = +2
Query: 510 GGSCPKGSECSKHVTAGVSIHS---SAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQ 566
G + +ECS+HV VS S SAE QRRFDTSSYQQRAEALEGLLEFSARLLQ Q
Sbjct: 29 GAAVSHSNECSEHVATAVSSRSHSSSAESCQRRFDTSSYQQRAEALEGLLEFSARLLQQQ 208
Query: 567 RFDELGVLLKPFGLEKVSPRETAIWLAKSFKETVI 601
RF+ELGVLLKPFG EKVSPRETAIWL KSFKETV+
Sbjct: 209 RFEELGVLLKPFGQEKVSPRETAIWLTKSFKETVV 313
>AV767547
Length = 288
Score = 129 bits (325), Expect = 1e-30
Identities = 64/66 (96%), Positives = 66/66 (99%)
Frame = -2
Query: 536 HQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS 595
HQRRFDTSSYQQRA+AL+GLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS
Sbjct: 287 HQRRFDTSSYQQRAKALKGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKS 108
Query: 596 FKETVI 601
FKETVI
Sbjct: 107 FKETVI 90
>BP055911
Length = 373
Score = 122 bits (306), Expect = 2e-28
Identities = 48/76 (63%), Positives = 67/76 (88%)
Frame = +2
Query: 56 SKVRNPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLM 115
+++++P+IVE+K++WVEKGCFVCI+ GYCE GD+A +KK+NG FPEEKLCKW+ QLL+
Sbjct: 68 ARIQHPYIVEFKEAWVEKGCFVCIVTGYCEGGDMAXLMKKSNGAYFPEEKLCKWITQLLL 247
Query: 116 ALDYLHVNHILHRDVK 131
A++YLH N +LHRD+K
Sbjct: 248 AVEYLHSNFVLHRDLK 295
Score = 37.7 bits (86), Expect = 0.005
Identities = 18/48 (37%), Positives = 30/48 (62%)
Frame = +3
Query: 34 KIRLARQSDRTRRSAHQEMELISKVRNPFIVEYKDSWVEKGCFVCIIV 81
KIRLARQ++R RRSA+QEM L+ + P ++ + + K +++
Sbjct: 3 KIRLARQTERCRRSAYQEMALMLEFSTPTLLNLRRHGLRKDVLSALLL 146
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 118 bits (295), Expect = 3e-27
Identities = 80/261 (30%), Positives = 128/261 (48%), Gaps = 7/261 (2%)
Frame = +2
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQE----MELISKV 58
QY++ E+IG+G FG+ H + +K I + +D T R + M L+S
Sbjct: 65 QYQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLLSP- 241
Query: 59 RNPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALD 118
+P I++ D + E + +++ C+ L D I ANG + PE + + QLL A+
Sbjct: 242 -HPNILQIFDVF-EDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVA 415
Query: 119 YLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLADI 178
+ H + HRDVK N+ D++L DFG A+ S +VGTP Y+ PE+L
Sbjct: 416 HCHRLGVAHRDVKPDNVLFGGGGDLKLADFGSAEWFGDGRRMSGVVGTPYYVAPEVLMGR 595
Query: 179 PYGSKSDIWSLGCCIYEMAALKPAF---KAFDIQALINKINKSIVAPLPTMYSSAFRGLV 235
YG K D+WS G +Y M + P F A +I + + N + + S A + L+
Sbjct: 596 EYGEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAVIRGNLRFPSRIFRNVSPAAKDLL 775
Query: 236 KSMLRKNPELRPPASELLNHP 256
+ M+ ++P R A + L HP
Sbjct: 776 RKMICRDPSNRISAEQALRHP 838
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 115 bits (287), Expect = 3e-26
Identities = 75/262 (28%), Positives = 136/262 (51%), Gaps = 5/262 (1%)
Frame = +3
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLAR-QSDRTRRSAHQEMELISKVRNP 61
+YE+ +G+G+F R+ + +K I+ + + DR + +E+ ++ VR+P
Sbjct: 429 KYEMGRVLGQGNFAKVYHGRNLATNENVAIKVIKKEKLKKDRLVKQIKREVSVMRLVRHP 608
Query: 62 FIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH 121
IVE K+ KG + +++ Y + G+L K N E+ K+ QL+ A+D+ H
Sbjct: 609 HIVELKEVMATKGK-IFLVMEYVKGGEL---FTKVNKGKLNEDDARKYFQQLISAVDFCH 776
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLGDFGLA---KMLTSDDLASSIVGTPSYMCPELLADI 178
+ HRD+K N+ L +N+D+++ DFGL+ + D + + GTP+Y+ PE+L
Sbjct: 777 SRGVTHRDLKPENLLLDENEDLKVSDFGLSALPEQRRDDGMLVTPCGTPAYVAPEVLKKK 956
Query: 179 PY-GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKS 237
Y GSK+DIWS G +Y + + F+ ++ + K K+ P S + L+ +
Sbjct: 957 GYDGSKADIWSCGVILYALLSGYLPFQGENVMRIYRKAFKA-EYEFPEWISPQAKNLISN 1133
Query: 238 MLRKNPELRPPASELLNHPHLQ 259
+L +PE R E+++ P Q
Sbjct: 1134LLVADPEKRYSIPEIISDPWFQ 1199
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 114 bits (286), Expect = 3e-26
Identities = 72/223 (32%), Positives = 119/223 (53%), Gaps = 2/223 (0%)
Frame = +3
Query: 39 RQSDRTRRSAHQEMELISKVRNPFIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANG 98
+ S + +QE+ L+++ +P IV+Y S + + + + + Y G + +++
Sbjct: 45 KTSKECLKQLNQEINLLNQFSHPNIVQYYGSELGEES-LSVYLEYVSGGSIHKLLQEYGA 221
Query: 99 VNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDD 158
F E + + Q++ L YLH + +HRD+K +NI + N +I+L DFG++K + S
Sbjct: 222 --FKEPVIQNYTRQIVSGLAYLHSRNTVHRDIKGANILVDPNGEIKLADFGMSKHINSAA 395
Query: 159 LASSIVGTPSYMCPELLADI-PYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINK 217
S G+P +M PE++ + YG DI SLGC I EMA KP + F+ A I KI
Sbjct: 396 SMLSFKGSPYWMAPEVVMNTNGYGLPVDISSLGCTILEMATSKPPWSQFEGVAAIFKIGN 575
Query: 218 SIVAP-LPTMYSSAFRGLVKSMLRKNPELRPPASELLNHPHLQ 259
S P +P S + +K L+++P RP A LLNHP ++
Sbjct: 576 SKDMPEIPEHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFIR 704
>TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein kinase Abl
(D-ash) , partial (5%)
Length = 1197
Score = 105 bits (261), Expect = 3e-23
Identities = 59/223 (26%), Positives = 119/223 (52%), Gaps = 1/223 (0%)
Frame = +2
Query: 32 LKKIRLARQSDRTRRSAHQEMELISKVRNPFIVEYKDSWVEKGCFVCIIVGYCERGDLAD 91
+K ++ R S + QE+ ++ K+R+ +V++ + +CI+ + RG L D
Sbjct: 23 IKVLKPERISTDMLKEFAQEVYIMRKIRHKNVVQFIGACTRTPN-LCIVTEFMSRGSLYD 199
Query: 92 AIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLA 151
+ K GV F L K + + ++YLH N+I+HRD+K N+ + +N+ +++ DFG+A
Sbjct: 200 FLHKQRGV-FKLPSLLKVAIDVSKGMNYLHQNNIIHRDLKTGNLLMDENELVKVADFGVA 376
Query: 152 KMLTSDDLASSIVGTPSYMCPELLADIPYGSKSDIWSLGCCIYEMAALK-PAFKAFDIQA 210
+++T + ++ GT +M PE++ PY K+D++S G ++E+ + P +QA
Sbjct: 377 RVITQSGVMTAETGTYRWMAPEVIEHKPYDQKADVFSFGIALWELLTGELPYSYLTPLQA 556
Query: 211 LINKINKSIVAPLPTMYSSAFRGLVKSMLRKNPELRPPASELL 253
+ + K + +P L++ +++P RP SE++
Sbjct: 557 AVGVVQKGLRPSIPKNTHPRLSELLQRCWKQDPIERPAFSEII 685
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 100 bits (250), Expect = 5e-22
Identities = 62/199 (31%), Positives = 108/199 (54%), Gaps = 5/199 (2%)
Frame = +3
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLAR-QSDRTRRSAHQEMELISKVRNP 61
+YE+ + +G+G+F R+ + +K I+ R + +R + +E+ ++ VR+P
Sbjct: 339 KYEIGKILGQGNFAKVYHGRNMETNESVAIKVIKKERLKKERLVKQIKREVSVMRLVRHP 518
Query: 62 FIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH 121
IVE K+ K + ++V Y + G+L + K E K+ QL+ A+D+ H
Sbjct: 519 HIVELKEVMATK-TKIFMVVEYVKGGELFAKLTKGK---MTEVAARKYFQQLISAVDFCH 686
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLGDFGLAKM---LTSDDLASSIVGTPSYMCPELLADI 178
+ HRD+K N+ L N+D+++ DFGL+ + SD + + GTP+Y+ PE+L
Sbjct: 687 SRGVTHRDLKPENLLLDDNEDLKVSDFGLSSLPEQRRSDGMLLTPCGTPAYVAPEVLKKK 866
Query: 179 PY-GSKSDIWSLGCCIYEM 196
Y GSK+DIWS G +Y +
Sbjct: 867 GYDGSKADIWSCGVILYAL 923
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 95.9 bits (237), Expect = 2e-20
Identities = 67/256 (26%), Positives = 125/256 (48%), Gaps = 5/256 (1%)
Frame = +2
Query: 3 QYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLAR-QSDRTRRSAHQEMELISKVRNP 61
+YE+ +GKG+ + + +K + AR + + +E+ ++ VR+P
Sbjct: 35 KYEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMRLVRHP 214
Query: 62 FIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH 121
IV K+ K + I+ Y G+L + K ++ ++ QL+ A+DY H
Sbjct: 215 NIVNLKEVMATK-TKIFFIMEYIRGGELFAKVAKGK---LKDDLARRYFQQLISAVDYCH 382
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLGDFGLA---KMLTSDDLASSIVGTPSYMCPELLADI 178
+ HRD+K N+ L +N+++++ DFGL+ + L D L + GTP+Y+ PE+L
Sbjct: 383 SRGVSHRDLKPENLLLDENENLKVSDFGLSGLPEQLRQDGLLHTQCGTPAYVAPEVLRKK 562
Query: 179 PY-GSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKS 237
Y G K+D WS G +Y + A F+ ++ + NK+ ++ P +S + L+
Sbjct: 563 GYDGFKTDTWSCGVILYALLAGCLPFQHENLMTMYNKVLRA-EFQFPPWFSPESKKLISK 739
Query: 238 MLRKNPELRPPASELL 253
+L +P R S ++
Sbjct: 740 ILVADPNRRITISSIM 787
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 94.0 bits (232), Expect = 6e-20
Identities = 68/253 (26%), Positives = 119/253 (46%), Gaps = 5/253 (1%)
Frame = +3
Query: 10 IGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNPFIVEYKDS 69
IG+G++G + + +L +KKI A + + +E++L+ + + ++ +D
Sbjct: 309 IGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLREIKLLRHLDHENVIALRDV 488
Query: 70 W---VEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHIL 126
+ + I DL I+ G++ EE +L Q+L L Y+H +I+
Sbjct: 489 IPPPLRREFTDVYITTELMDTDLHQIIRSNQGLS--EEHCQYFLYQVLRGLKYIHSANII 662
Query: 127 HRDVKCSNIFLTKNQDIRLGDFGLAKMLTSDDLASSIVGTPSYMCPELLAD-IPYGSKSD 185
HRD+K SN+ L N D+++ DFGLA+ +D + V T Y PELL + Y S D
Sbjct: 663 HRDLKPSNLLLNSNCDLKIIDFGLARPTVENDFMTEYVVTRWYRAPELLLNSSDYTSAID 842
Query: 186 IWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAPLPTMYSSAFRGLVKSMLRKNPEL 245
+WS+GC E+ KP F D + + + + P V+ +R+ P+
Sbjct: 843 VWSVGCIFMELMNKKPLFPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVRRYIRQLPQY 1022
Query: 246 -RPPASELLNHPH 257
R P +++ H H
Sbjct: 1023PRQPLTKVFPHVH 1061
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 93.6 bits (231), Expect = 8e-20
Identities = 62/192 (32%), Positives = 99/192 (51%), Gaps = 3/192 (1%)
Frame = +3
Query: 2 EQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNP 61
E+YE L+++G G+FG A L R K+ +L +K I ++ D + +E+ +R+P
Sbjct: 72 ERYEPLKELGSGNFGVARLARDKNTGELVAVKYIERGKKID---ENVQREIINHRSLRHP 242
Query: 62 FIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH 121
I+ +K+ + + I++ Y G+L + I A F E++ + QL+ + Y H
Sbjct: 243 NIIRFKEVLLTP-THLAIVLEYASGGELFERICSAG--RFSEDEARYFFQQLISGVSYCH 413
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLG--DFGLAKMLTSDDLASSIVGTPSYMCPELLADIP 179
I HRD+K N L N RL DFG +K S VGTP+Y+ PE+L+
Sbjct: 414 SMEICHRDLKLENTLLDGNPSPRLKICDFGYSKSAILHSQPKSTVGTPAYIAPEVLSRKE 593
Query: 180 Y-GSKSDIWSLG 190
Y G +D+WS G
Sbjct: 594 YDGKVADVWSCG 629
>TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated protein
kinase homolog MMK1 (MAP kinase MSK7) (MAP kinase ERK1)
, partial (71%)
Length = 1098
Score = 88.2 bits (217), Expect = 3e-18
Identities = 62/201 (30%), Positives = 93/201 (45%), Gaps = 30/201 (14%)
Frame = +1
Query: 88 DLADAIKKANGVNFPEEKLCKWLVQLLMALDYLHVNHILHRDVKCSNIFLTKNQDIRLGD 147
DL I+ G++ EE +L Q+L L Y+H ++LHRD+K SN+ L N D+++ D
Sbjct: 94 DLHQIIRSNQGLS--EEHCQYFLYQILRGLKYIHSANVLHRDLKPSNLLLNANCDLKICD 267
Query: 148 FGLAKMLTSDDLASSIVGTPSYMCPELLAD-IPYGSKSDIWSLGCCIYEMAALKPAF--- 203
FGLA++ + D + V T Y PELL + Y + D+WS+GC E+ KP F
Sbjct: 268 FGLARVTSETDFMTEYVVTRWYRAPELLLNSSDYTAAIDVWSVGCIFMELMDRKPLFPGR 447
Query: 204 ---------------KAFDIQALINKINKSIVAPLPTMYSSAFR-----------GLVKS 237
+ D +N+ K + LP +F+ LV+
Sbjct: 448 DHVHQLRLLMELIGTPSEDDLGFLNENAKRYIRQLPPYRRQSFQEKFPQVHPEAIDLVEK 627
Query: 238 MLRKNPELRPPASELLNHPHL 258
ML +P R E L HP+L
Sbjct: 628 MLTFDPRKRITVEEALAHPYL 690
>AV769753
Length = 496
Score = 85.5 bits (210), Expect = 2e-17
Identities = 46/80 (57%), Positives = 56/80 (69%)
Frame = -3
Query: 518 ECSKHVTAGVSIHSSAELHQRRFDTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKP 577
E SK +T + L + D S++QRAEALE LLEFSA LLQ +R +EL V+LKP
Sbjct: 485 ELSKCITP--TTEDKPPLAKETMDVKSFRQRAEALEELLEFSAELLQQERLEELKVVLKP 312
Query: 578 FGLEKVSPRETAIWLAKSFK 597
FG +KVSPRETAIWLA+S K
Sbjct: 311 FGKDKVSPRETAIWLARSLK 252
>TC15873 similar to UP|Q9LXP3 (Q9LXP3) Protein kinase-like protein, partial
(7%)
Length = 522
Score = 82.4 bits (202), Expect = 2e-16
Identities = 41/61 (67%), Positives = 47/61 (76%)
Frame = +1
Query: 541 DTSSYQQRAEALEGLLEFSARLLQHQRFDELGVLLKPFGLEKVSPRETAIWLAKSFKETV 600
D S QRAEALEGLLE SA LLQ R +EL ++LKPFG +KVSPRETAIWLAKS K +
Sbjct: 67 DVKSSTQRAEALEGLLELSADLLQQNRLEELSIVLKPFGKDKVSPRETAIWLAKSLKGLM 246
Query: 601 I 601
+
Sbjct: 247 V 249
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 82.0 bits (201), Expect = 2e-16
Identities = 53/177 (29%), Positives = 91/177 (50%), Gaps = 2/177 (1%)
Frame = +1
Query: 2 EQYEVLEQIGKGSFGSALLVRHKHEKKLYVLKKIRLARQSDRTRRSAHQEMELISKVRNP 61
++Y+++ IG G+FG A L++ K K+L +K I + D+ + +E+ +R+P
Sbjct: 355 DRYDLVRDIGSGNFGVARLMQDKQTKELVAVKYIE---RGDKIDENVKREIINHRSLRHP 525
Query: 62 FIVEYKDSWVEKGCFVCIIVGYCERGDLADAIKKANGVNFPEEKLCKWLVQLLMALDYLH 121
IV +K+ + + I++ Y G+L + I N F E++ + QL+ + Y H
Sbjct: 526 NIVRFKEV-ILTPTHLAIVMEYASGGELFERI--CNAGRFTEDEARFFFQQLISGVSYCH 696
Query: 122 VNHILHRDVKCSNIFLTKNQDIRLG--DFGLAKMLTSDDLASSIVGTPSYMCPELLA 176
+ HRD+K N L + RL DFG +K S VGTP+Y+ PE+L+
Sbjct: 697 AMQVCHRDLKLENTLLDGSPTPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLS 867
>TC15973 homologue to GB|AAL15278.1|16323087|AY057647 AT3g01490/F4P13_4
{Arabidopsis thaliana;}, partial (49%)
Length = 944
Score = 80.1 bits (196), Expect = 9e-16
Identities = 43/147 (29%), Positives = 75/147 (50%), Gaps = 3/147 (2%)
Frame = +3
Query: 111 VQLLMALDYLHVNHILHRDVKCSNIFLTKNQDIRLGDFGLAKMLTS--DDLASSIVGTPS 168
+ L L YLH I+HRDVK N+ L K + +++ DFG+A++ S +D+ GT
Sbjct: 78 LDLARGLSYLHSQKIVHRDVKTENMLLDKTRTVKIADFGVARVEASNPNDMTGE-TGTLG 254
Query: 169 YMCPELLADIPYGSKSDIWSLGCCIYEMAALKPAFKAFDIQALINKINKSIVAP-LPTMY 227
YM PE+L PY K D++S G C++E+ + + + + + + P +P
Sbjct: 255 YMAPEVLNGNPYNRKCDVYSFGICLWEIYCCDMPYPDLSFSEITSAVVRQNLRPEIPRCC 434
Query: 228 SSAFRGLVKSMLRKNPELRPPASELLN 254
S+ ++K P+ RP E+++
Sbjct: 435 PSSLANVMKKCWDATPDKRPEMDEVVS 515
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,294,742
Number of Sequences: 28460
Number of extensions: 170285
Number of successful extensions: 1454
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 1387
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1405
length of query: 601
length of database: 4,897,600
effective HSP length: 95
effective length of query: 506
effective length of database: 2,193,900
effective search space: 1110113400
effective search space used: 1110113400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0322.12


