

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.5
(1411 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
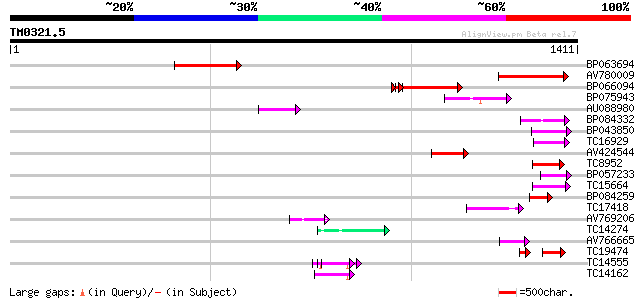
Score E
Sequences producing significant alignments: (bits) Value
BP063694 190 2e-48
AV780009 159 2e-39
BP066094 135 6e-32
BP075943 105 4e-23
AU088980 82 8e-16
BP084332 77 2e-14
BP043850 75 5e-14
TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%) 70 2e-12
AV424544 69 4e-12
TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%) 68 1e-11
BP057233 65 6e-11
TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, p... 64 2e-10
BP084259 59 4e-09
TC17418 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partia... 54 1e-07
AV769206 54 2e-07
TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.3... 53 3e-07
AV766665 52 6e-07
TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retro... 37 8e-07
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 52 8e-07
TC14162 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein p... 51 1e-06
>BP063694
Length = 511
Score = 190 bits (482), Expect = 2e-48
Identities = 110/168 (65%), Positives = 125/168 (73%)
Frame = -1
Query: 410 YFSG*FFCSSEPTDSSRSGKGPL**GTLCA*SRIAGAAYYQFSITSCLL*TVALPLRTC* 469
+F G*F C SE D RSGKG +* TLCA S +AG AYYQFSIT CL * VAL +RT
Sbjct: 511 HFFG*FXCDSESKDRRRSGKGQM*SRTLCAESGLAGVAYYQFSITPCLF*IVALKIRTRQ 332
Query: 470 F*NYSTIA*TWLFQCFLNFS*AYLLYFLSEG*E*TSSLL****KSFCSFRPYPL*FVGTI 529
F* Y TIA*TWLF+CFL F+*A+LL+FLS+G*+*T S+ **** SFCSFRPY L* GT+
Sbjct: 331 F*YYQTIA*TWLFRCFLYFA*AHLLHFLSDG*K*TISIS****ASFCSFRPYSL*LEGTV 152
Query: 530 SCCFC**FFVLCYFCG*FFPVYLVLSVEAQDWFL*CSCSLQSVCGKSI 577
S CF * F+LCY C * P YLVLS+EA F *C+ SLQSV GKSI
Sbjct: 151 SRCFY*WVFLLCYIC**LLPFYLVLSIEA*V*FF*CTSSLQSVYGKSI 8
>AV780009
Length = 529
Score = 159 bits (403), Expect = 2e-39
Identities = 84/176 (47%), Positives = 108/176 (60%)
Frame = -1
Query: 1216 QAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSW 1275
QA R+LRYV G GL F S + YSD+DWA C DTRRS GY+IFLG +L+SW
Sbjct: 529 QAATRVLRYVKGAPAQGLFFSADSPLKLQAYSDSDWAGCPDTRRSVTGYSIFLGTSLISW 350
Query: 1276 SAKKQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQN 1335
KKQ TV+RSS E+EYRA+A T E+ WL L L++ + L DNQSAL +A N
Sbjct: 349 RTKKQTTVSRSSSEAEYRALAATVCEVQWLSYLFQFLKLNVPLPVPLFCDNQSALHIAHN 170
Query: 1336 PVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFRSK 1391
P H+ KHI+LDCH V + +G + + + T QLADIFTK P F + ++
Sbjct: 169 PTFHERTKHIELDCHVVRAKLQAGLIHLLPISTHHQLADIFTKPCPHLCFPLLGAR 2
>BP066094
Length = 532
Score = 135 bits (339), Expect(3) = 6e-32
Identities = 66/148 (44%), Positives = 100/148 (66%)
Frame = +2
Query: 978 KATTVRLILSHVVLNDWQLHQLDVKNAFLHGHLTETVYMEQPPGFVDPRFPTHVCRLNKA 1037
K +RL++S V ++ LHQ++VK+AFL+G+++E VY+ QPPG D + H+ +L K+
Sbjct: 89 KTEAIRLLISFSVNHNIILHQMNVKSAFLNGYISEEVYVHQPPGXEDEKNSDHIFKLKKS 268
Query: 1038 LYGLKQAPRAWFQRLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPS 1097
LYGLKQAPRAW++RLSSFLL + + D +LF L + +YVDDII ++PS
Sbjct: 269 LYGLKQAPRAWYERLSSFLLENEXVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANPS 448
Query: 1098 LLTQFIARLNAEFAIKYLGKLGYFLGLE 1125
L +F + AEF ++ +G+L YFLG++
Sbjct: 449 LCKEFSEMMQAEFEMRMMGELKYFLGIQ 532
Score = 21.6 bits (44), Expect(3) = 6e-32
Identities = 8/18 (44%), Positives = 13/18 (71%)
Frame = +3
Query: 961 FTQIPGFDYSLTFSPVVK 978
++Q G DY+ TF+PV +
Sbjct: 39 YSQQ*GIDYTETFAPVAR 92
Score = 19.6 bits (39), Expect(3) = 6e-32
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = +1
Query: 951 RLKARLVAQG 960
R KARLVAQG
Sbjct: 10 RNKARLVAQG 39
>BP075943
Length = 547
Score = 105 bits (263), Expect = 4e-23
Identities = 69/178 (38%), Positives = 98/178 (54%), Gaps = 11/178 (6%)
Frame = -3
Query: 1082 LLVYVDDIILTGSDPSLLTQFIARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQAKYA 1141
LL+YVDD++L G+D + + L+ +F IK LG+ YFLGLEI + G+ L Q KYA
Sbjct: 521 LLLYVDDVLLAGNDMHEIQLVKSSLHDQFRIKDLGEAKYFLGLEIARSTSGIVLNQRKYA 342
Query: 1142 HDLLSRAMMLEASHVSTPLAAGSHLVSS-------GEAYFDPTHYRSLVGALQYLTITRP 1194
L+S ++ H SH +S G D YR +VG L YL TRP
Sbjct: 341 LQLIS-----DSGHFGFQPRFYSHGTNSQTLGTNTGTPLTDIGSYRRIVGRLLYLNTTRP 177
Query: 1195 DLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSS----PAVLGYSD 1248
D+++AVN +SQFL APT H Q + L+Y+ G+ GL + +SS P+++G D
Sbjct: 176 DITFAVNQLSQFLSAPTDIHEQQLTGFLKYL*GSPGSGLFYPASSSTL*LPSLIGGPD 3
>AU088980
Length = 360
Score = 81.6 bits (200), Expect = 8e-16
Identities = 36/105 (34%), Positives = 58/105 (54%)
Frame = +2
Query: 619 NGRVEQKHRHVLELGLAMLYHSHVPTRYWVDAFSTAVYIINRVPSKVLSNQIPFQLLFHV 678
N VE+KHRH+L + A+++HS++P W A A Y+IN +PS +L Q P+Q L +
Sbjct: 2 NSIVERKHRHILNVTRALMFHSYLPKNLWTFAVKHAAYLINXLPSPLLKGQCPYQFLNND 181
Query: 679 APTYANFHPFGCRVFPCLRPYMNNKFSPRSTPCIFFGYSSHHKGF 723
+PT + FG + + K PR+ C+F G+ + G+
Sbjct: 182 SPTLLDLKVFGTLCYSSTLTHNXQKLDPRARKCVFLGFKTGTXGY 316
>BP084332
Length = 368
Score = 77.0 bits (188), Expect = 2e-14
Identities = 42/123 (34%), Positives = 70/123 (56%), Gaps = 1/123 (0%)
Frame = +2
Query: 1271 NLLSWSAK-KQPTVARSSCESEYRAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSA 1329
N+ W A Q T+A S+ E+EY + A +++++W+ + L + ++ S P+ DN +A
Sbjct: 2 NVNFWEAT*GQSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQILESNIPIYC-DNTAA 178
Query: 1330 LFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRPLFEIFR 1389
+ +++NP+ H AKHI++ HF+ + V G L ++ V T Q ADIFTK L F
Sbjct: 179 ISLSKNPILHSRAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFIL 358
Query: 1390 SKL 1392
L
Sbjct: 359 KNL 367
>BP043850
Length = 515
Score = 75.5 bits (184), Expect = 5e-14
Identities = 42/99 (42%), Positives = 58/99 (58%)
Frame = -1
Query: 1300 SELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASG 1359
SE++WL LL EL S L +DN SA+ +A NPV H+ +HI++DCH V E
Sbjct: 515 SEIIWLRGLLSELGFLQSQPTPLHADNTSAIQIAANPVYHEWTRHIEVDCHSVREAYDRR 336
Query: 1360 RLAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRVGLNP 1398
+ + HV TS+Q+ADI TK+L R SKL + +P
Sbjct: 335 VITLPHVSTSVQIADILTKSLTRQRHNFLVSKLMLVESP 219
>TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%)
Length = 553
Score = 70.5 bits (171), Expect = 2e-12
Identities = 37/89 (41%), Positives = 51/89 (56%)
Frame = +1
Query: 1304 WLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAV 1363
WL LL +L+V L+ DN SA +A NPV H+ KHI++DCH V E + G + +
Sbjct: 4 WLTYLLQDLKVPFEQPALVYCDNNSARHIAANPVFHERTKHIEIDCHIVRERIQKGLIHL 183
Query: 1364 RHVPTSLQLADIFTKALPRPLFEIFRSKL 1392
+ +S LADI+TKAL F +KL
Sbjct: 184 LPISSSEPLADIYTKALSPQNFHQICAKL 270
>AV424544
Length = 276
Score = 69.3 bits (168), Expect = 4e-12
Identities = 40/90 (44%), Positives = 56/90 (61%)
Frame = +3
Query: 1051 RLSSFLLRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFIARLNAEF 1110
+LSS+L G+ S D SLF ++ + +LVYVDD+IL G+D + + +L+ +F
Sbjct: 6 KLSSYLHILGYIQSAHDHSLFTKFRDASFTVILVYVDDLILAGNDLNEIQCVKNKLDIQF 185
Query: 1111 AIKYLGKLGYFLGLEITYTPDGLFLGQAKY 1140
IK LG L YFLGLE+ + GLFL Q KY
Sbjct: 186 RIKDLGTLKYFLGLEVARSSCGLFLSQRKY 275
>TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%)
Length = 550
Score = 67.8 bits (164), Expect = 1e-11
Identities = 34/80 (42%), Positives = 50/80 (62%)
Frame = +3
Query: 1301 ELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGR 1360
E +WL L +LR+ ++ DN+SAL +A N V HK ++I++DCH V V G
Sbjct: 21 EALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFGI 200
Query: 1361 LAVRHVPTSLQLADIFTKAL 1380
L + HVP+S Q+AD+FTK +
Sbjct: 201 LHLLHVPSSDQVADVFTKTI 260
>BP057233
Length = 473
Score = 65.5 bits (158), Expect = 6e-11
Identities = 33/79 (41%), Positives = 46/79 (57%)
Frame = -2
Query: 1320 PLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKA 1379
P+L DN SA +A NPV H +KHI++D H++ + V + V +VPT+ Q+AD TK
Sbjct: 451 PILWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNEVVVAYVPTTDQIADCLTKP 272
Query: 1380 LPRPLFEIFRSKLRVGLNP 1398
L F R KL V +P
Sbjct: 271 LSHTRFSQLRDKLGVIHSP 215
>TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, partial (4%)
Length = 670
Score = 63.5 bits (153), Expect = 2e-10
Identities = 38/94 (40%), Positives = 51/94 (53%)
Frame = +1
Query: 1301 ELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGR 1360
EL WL +L +LRV + LL D+QSA +A N V H+ KH+D+DCH V E + +
Sbjct: 4 EL*WLTYILQDLRVPFISPSLLYCDSQSARHIATNAVFHERTKHLDIDCHVVREKLQAKL 183
Query: 1361 LAVRHVPTSLQLADIFTKALPRPLFEIFRSKLRV 1394
+ + + Q ADI TK L F SKL V
Sbjct: 184 FHLLPISSVDQTADILTKPLESGPFSHLVSKLGV 285
>BP084259
Length = 385
Score = 59.3 bits (142), Expect = 4e-09
Identities = 26/59 (44%), Positives = 37/59 (62%)
Frame = -3
Query: 1293 RAMANTASELVWLLNLLHELRVRLSATPLLLSDNQSALFMAQNPVAHKHAKHIDLDCHF 1351
R + +TA+E +WL LL EL+ L P + S+N +A ++ N V H H KH+ LDCHF
Sbjct: 179 RTIGSTAAEFLWLQQLLKELQFSLPKVPCIFSNNINATYVCLNLVFHSHMKHVALDCHF 3
>TC17418 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partial (20%)
Length = 739
Score = 54.3 bits (129), Expect = 1e-07
Identities = 44/145 (30%), Positives = 70/145 (47%), Gaps = 4/145 (2%)
Frame = -1
Query: 1137 QAKYAHDLLSRAMMLEASHVSTPLAAGSHLVS---SGEAYFDPTHYRSLVGALQYLTITR 1193
Q Y D+L + M + ++ST + G + + +G D T+Y+SL+G+++YL R
Sbjct: 739 QKIYVDDILKKFKMTNSKYISTTIG-GKEIEAGRRNGGKRVDSTYYKSLIGSVRYLNTVR 563
Query: 1194 PDLSYAVNTVSQFLQAPTMEHFQAVKRILRYVCGTQHFGLTFRRTSSPAVLGYSDADWAR 1253
D+ V S+F++ P H Q +R LRY+ GT G+ + L
Sbjct: 562 SDIVCGVGLRSRFME-P*DCH*QGAQRSLRYIKGTLKDGIFMLVITM*NSL--------- 413
Query: 1254 CTDTRRSTY-GYAIFLGDNLLSWSA 1277
DT+ T GYA LG +SWS+
Sbjct: 412 --DTQIVTRPGYAFHLGRGAISWSS 344
>AV769206
Length = 536
Score = 53.5 bits (127), Expect = 2e-07
Identities = 36/103 (34%), Positives = 48/103 (45%), Gaps = 3/103 (2%)
Frame = -3
Query: 696 LRPYMNNKFSPRSTPCIFFGYSSHHKGFKCFNPAISLTYVSHHAQFDEFCFPFAGSHSSP 755
LRPY + K S S C+F GYS +HKG+KC + + Y+S F E FP+
Sbjct: 531 LRPYNSTKLSLHSKECVFIGYSINHKGYKCLDQS-GRIYISKDVLFHEHRFPYPSLFPDE 355
Query: 756 HDLVVSTFYEPASGSPPSPH---PVVLVAPESSLPIGSLSCPS 795
S + P S P H P + + SSL GS + PS
Sbjct: 354 TTSSTSAEFFPLSTIPIVSHTTPPALASSGFSSL--GSATLPS 232
>TC14274 weakly similar to GB|AAF79559.1|8778551|AC022464 F22G5.35
{Arabidopsis thaliana;} , partial (16%)
Length = 1208
Score = 53.1 bits (126), Expect = 3e-07
Identities = 49/182 (26%), Positives = 71/182 (38%), Gaps = 3/182 (1%)
Frame = +3
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
P + SPP+P P + P G+ PS + L + P P PP+P +
Sbjct: 168 PTAVSPPNP-----TTPATPPPSGTSVSPSLSLTPSLTPFSPSRSSTPNPPIPPNP---S 323
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRSQN---GIFRPNPRYALVHAQPTGI 882
+PP + +P ++ PP A P + A P RS + + PNP A PT
Sbjct: 324 SPPSASSSPTSTTSMIPPASANSPSSAPPAAPTARSTSSSASLVAPNPSITPTQA-PTPT 500
Query: 883 LTAFHTVTKPKGFKSAMKHP*WLAAMEDELSALHKNCTWTLVPRPSTTNVVGIKWVFRTK 942
LT TVT + H L + L L L+P P+ T V I+ +
Sbjct: 501 LTLTLTVTLRSSITEDILH--HLPNLNHLLLTLLPTPLTLLIPTPAVTIPVTIQVLLHRH 674
Query: 943 FH 944
H
Sbjct: 675 HH 680
>AV766665
Length = 601
Score = 52.0 bits (123), Expect = 6e-07
Identities = 28/75 (37%), Positives = 43/75 (57%)
Frame = +3
Query: 1220 RILRYVCGTQHFGLTFRRTSSPAVLGYSDADWARCTDTRRSTYGYAIFLGDNLLSWSAKK 1279
RI Y+ G F++ ++ G++ AD+ R ST GY +FL NL++W +K+
Sbjct: 372 RIF*YLKANSRRGPLFQKEGKSSMDGFTYADYLGSIVDRLSTMGYYMFLSGNLVTWRSKQ 551
Query: 1280 QPTVARSSCESEYRA 1294
Q +ARSS E+E RA
Sbjct: 552 QNIIARSSGEAELRA 596
Score = 32.0 bits (71), Expect = 0.70
Identities = 26/94 (27%), Positives = 43/94 (45%), Gaps = 3/94 (3%)
Frame = +1
Query: 1047 AWFQRLSSFL---LRHGFSCSRADPSLFFFYKGHTTLYLLVYVDDIILTGSDPSLLTQFI 1103
AWF RL+ R +S ++ P + HT+ +L VYVD + + G D
Sbjct: 13 AWFVRLTEATHAESRWPYSLNKTCPQR----QTHTSSHLCVYVDHMTIVGDDEVEKQILR 180
Query: 1104 ARLNAEFAIKYLGKLGYFLGLEITYTPDGLFLGQ 1137
+L +F +K KL Y +E+ Y +F+ +
Sbjct: 181 EKLAPQFDLK---KLKYISVIEVVYP**NIFISK 273
>TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retroelement pol
polyprotein, partial (3%)
Length = 517
Score = 36.6 bits (83), Expect(2) = 8e-07
Identities = 16/29 (55%), Positives = 21/29 (72%)
Frame = -1
Query: 1268 LGDNLLSWSAKKQPTVARSSCESEYRAMA 1296
L DNL+SW +K+ VARS E+E+R MA
Sbjct: 517 LSDNLISWKSKETIIVARSRAEAEFRVMA 431
Score = 34.7 bits (78), Expect(2) = 8e-07
Identities = 21/59 (35%), Positives = 36/59 (60%)
Frame = -2
Query: 1325 DNQSALFMAQNPVAHKHAKHIDLDCHFVCELVASGRLAVRHVPTSLQLADIFTKALPRP 1383
DN++AL M + + + AKHI++D HF+ + ++ R V ++ QL D+FTK+ P
Sbjct: 354 DNKAALHMPKI*FSMR-AKHIEIDFHFL*DRRLYLDISTRFVNSNDQLTDVFTKSWRGP 181
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 51.6 bits (122), Expect = 8e-07
Identities = 33/92 (35%), Positives = 42/92 (44%)
Frame = +2
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
PA +P +P P A +SS P S P S PPA PP PA SP +
Sbjct: 215 PAPPTPTTPQPP---AAQSSPPPAQSSPPPVQSSP-------PPASTPP-PAQSSPPPVS 361
Query: 826 TPPVGTQAPPASPPTTPPVVAQVPLAPIAARP 857
+PP PP +P +TPP + P +P A P
Sbjct: 362 SPPPVQSTPPPAPASTPPPASPPPFSPPPATP 457
Score = 47.0 bits (110), Expect = 2e-05
Identities = 45/133 (33%), Positives = 58/133 (42%), Gaps = 10/133 (7%)
Frame = +2
Query: 753 SSPHDLVVSTFYEPASGSPP-----SPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPV 807
+SP +T PA+ S P SP PV P +S P + S P S P
Sbjct: 209 TSPAPPTPTTPQPPAAQSSPPPAQSSPPPVQSSPPPASTPPPAQSSPPPVSS------PP 370
Query: 808 PPADAPPSPAPPSPTQTATPPVGTQAPPASPP----TTPPVVAQVP-LAPIAARPRTRSQ 862
P PP PAP S A+PP PPA+PP T PP + VP +P A + +S+
Sbjct: 371 PVQSTPP-PAPASTPPPASPP-PFSPPPATPPPPAATPPPALTPVPATSPAPAPAKVKSK 544
Query: 863 NGIFRPNPRYALV 875
+ P P A V
Sbjct: 545 SPAPAPAPALAPV 583
Score = 44.3 bits (103), Expect = 1e-04
Identities = 32/94 (34%), Positives = 43/94 (45%), Gaps = 13/94 (13%)
Frame = +2
Query: 777 VVLVAPESSLPIGSLSCPSCDDSDVL-PALPVPPADAPPSP-APPSPTQTATPPVGTQAP 834
VVLV + I S+ S +S PA P P PP+ + P P Q++ PPV + P
Sbjct: 134 VVLVLGLICIVIASVGAQSPSNSPTTSPAPPTPTTPQPPAAQSSPPPAQSSPPPVQSSPP 313
Query: 835 PASPP-----------TTPPVVAQVPLAPIAARP 857
PAS P + PPV + P AP + P
Sbjct: 314 PASTPPPAQSSPPPVSSPPPVQSTPPPAPASTPP 415
Score = 38.1 bits (87), Expect = 0.010
Identities = 21/54 (38%), Positives = 31/54 (56%), Gaps = 1/54 (1%)
Frame = +2
Query: 809 PADAPP-SPAPPSPTQTATPPVGTQAPPASPPTTPPVVAQVPLAPIAARPRTRS 861
P+++P SPAPP+PT T PP +PP + + PPV + P P + P +S
Sbjct: 191 PSNSPTTSPAPPTPT-TPQPPAAQSSPPPAQSSPPPVQSSPP--PASTPPPAQS 343
Score = 37.4 bits (85), Expect = 0.017
Identities = 32/95 (33%), Positives = 39/95 (40%), Gaps = 5/95 (5%)
Frame = +2
Query: 766 PASGSPP---SPHPVVLVAP-ESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSP 821
PAS PP SP PV P +S+ P S P + P P P PP+ PP P
Sbjct: 314 PASTPPPAQSSPPPVSSPPPVQSTPPPAPASTPP--PASPPPFSPPPATPPPPAATPP-P 484
Query: 822 TQTATPPVGTQAPPASPPTTPPVVAQVP-LAPIAA 855
T P PA + P A P LAP+ +
Sbjct: 485 ALTPVPATSPAPAPAKVKSKSPAPAPAPALAPVVS 589
Score = 31.6 bits (70), Expect = 0.91
Identities = 28/88 (31%), Positives = 33/88 (36%), Gaps = 4/88 (4%)
Frame = +2
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPSPAPPSPTQTA 825
PA S P P P S P PA P PPA PP P P +
Sbjct: 392 PAPASTPPP----ASPPPFSPP---------------PATPPPPAATPPPALTPVPATSP 514
Query: 826 TP---PVGTQAP-PASPPTTPPVVAQVP 849
P V +++P PA P PVV+ P
Sbjct: 515 APAPAKVKSKSPAPAPAPALAPVVSLSP 598
>TC14162 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein precursor,
partial (87%)
Length = 1232
Score = 50.8 bits (120), Expect = 1e-06
Identities = 40/110 (36%), Positives = 47/110 (42%), Gaps = 10/110 (9%)
Frame = +2
Query: 758 LVVSTFYEPASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADA---PP 814
L++ F PAS PSP L P S + + PS S PA PA A P
Sbjct: 35 LLLLHFSYPASSFSPSPPTSHLHMPSS*PSSTTANTPSGPASSPTPATQSSPAAASSSTP 214
Query: 815 SPAPPSPTQTATPPVGTQAPPASP-------PTTPPVVAQVPLAPIAARP 857
SP PSP+QT T P + PA+P P P A V AP A P
Sbjct: 215 SPTSPSPSQTLTGPAASGPAPAAPTPPATASPAHPATAAAVSSAPAPAVP 364
Score = 38.1 bits (87), Expect = 0.010
Identities = 29/87 (33%), Positives = 38/87 (43%), Gaps = 12/87 (13%)
Frame = +2
Query: 766 PASGSPPSPHPVVLVAPESSLPIGSLSCPSCDDSDVLPALPVPPADAPPS--------PA 817
PA+ S P+ +P S P +L+ P+ S PA P PPA A P+ +
Sbjct: 170 PATQSSPAAASSSTPSPTSPSPSQTLTGPAA--SGPAPAAPTPPATASPAHPATAAAVSS 343
Query: 818 PPSPTQTATPP----VGTQAPPASPPT 840
P+P PP T A P SPPT
Sbjct: 344 APAPAVPPQPP*PSSTCTTATPTSPPT 424
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.347 0.151 0.556
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,781,853
Number of Sequences: 28460
Number of extensions: 676155
Number of successful extensions: 20040
Number of sequences better than 10.0: 837
Number of HSP's better than 10.0 without gapping: 9850
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 14312
length of query: 1411
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1309
effective length of database: 1,994,680
effective search space: 2611036120
effective search space used: 2611036120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0321.5


