

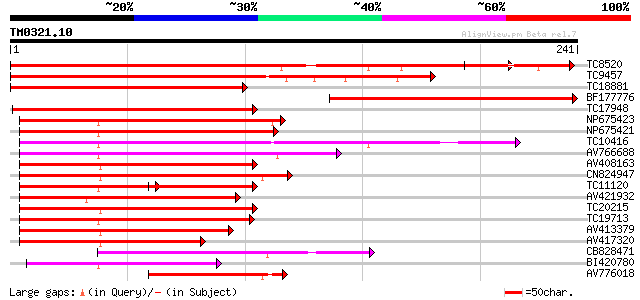
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0321.10
(241 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 240 1e-64
TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 238 7e-64
TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose... 214 1e-56
BF177776 200 2e-52
TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose... 175 7e-45
NP675423 transcription factor MYB101 [Lotus japonicus] 116 4e-27
NP675421 transcription factor MYB103 [Lotus japonicus] 115 5e-27
TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial... 114 1e-26
AV766688 108 6e-25
AV408163 106 4e-24
CN824947 105 5e-24
TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete 70 2e-23
AV421932 102 5e-23
TC20215 similar to AAS10104 (AAS10104) MYB transcription factor,... 102 6e-23
TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%) 102 8e-23
AV413379 95 1e-20
AV417320 84 2e-17
CB828471 79 5e-16
BI420780 68 1e-12
AV776018 55 8e-09
>TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (36%)
Length = 1778
Score = 240 bits (613), Expect = 1e-64
Identities = 129/233 (55%), Positives = 155/233 (66%), Gaps = 19/233 (8%)
Frame = +1
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDR+KGPWSPEEDEAL +LV+ HGP+NW+ ISKSIPGRSGKSCRLRWCNQLSP+VEHR F
Sbjct: 172 MDRIKGPWSPEEDEALQKLVERHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPQVEHRAF 351
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVD------ 114
TPEED IIRAHARFGNKWATIAR+L+GRTDNA+KNHWNSTLKRKCS++ A D
Sbjct: 352 TPEEDDTIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRKCSSIFAADDLAAGGG 531
Query: 115 -----HRPLKRSASVGAGCLNPSSPTGSAMSDPGPPAAISTD---APIAVPAAVVVPAT- 165
+PLKRSAS G P SP S +SD G A+S P+ V V ++
Sbjct: 532 GGDFCPQPLKRSASAGL----PGSPAWSDVSDSGCYVAVSPSHVFRPVPVRPYVETASSC 699
Query: 166 ----DPATSLSLSLPGFDSSGSGSGSKQLFGAEFLAVMQEMIRKEVRSYMSGM 214
P TSLSLSLPG DS+ + + + A ++ +V + G+
Sbjct: 700 EDDDGPPTSLSLSLPGVDSAEVTNRAAPVMAAPICPAPVTVVPAQVTAAQEGV 858
Score = 48.5 bits (114), Expect = 1e-06
Identities = 26/48 (54%), Positives = 32/48 (66%), Gaps = 1/48 (2%)
Frame = +2
Query: 194 EFLAVMQEMIRKEVRSYMSGMEEKNGACMP-AEAIGNAVMKRMGISNV 240
E +AVM EMIRKEVRSYM +++NG C + N +KRMGIS V
Sbjct: 884 ELMAVMHEMIRKEVRSYME--QQQNGVCFQGVDGFRNVAVKRMGISRV 1021
>TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (43%)
Length = 860
Score = 238 bits (607), Expect = 7e-64
Identities = 126/206 (61%), Positives = 144/206 (69%), Gaps = 25/206 (12%)
Frame = +3
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDR+KGPWSPEEDEAL RLV+ HGP+NW+ ISKSIPGRSGKSCRLRWCNQLSP+VEHR F
Sbjct: 66 MDRIKGPWSPEEDEALQRLVEKHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPQVEHRAF 245
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHR--PL 118
T EED IIRAHARFGNKWATIAR+L+GRTDNA+KNHWNSTLKRKC++ DH PL
Sbjct: 246 TAEEDDTIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRKCASF-MMDDHNAPPL 422
Query: 119 KRSASVGAGC----------LNPSSPTGSAMSD-------PGPPAAISTDAPIAVPAAVV 161
KRS S GA +P SP+GS +S+ P AA P+ V+
Sbjct: 423 KRSVSAGAATPVSTGLYMNPPSPGSPSGSDVSESSVPVCTPSAAAAAHVFRPVPRTGGVL 602
Query: 162 VP------ATDPATSLSLSLPGFDSS 181
P + DP TSLSLSLPG DS+
Sbjct: 603 PPVETTSSSNDPPTSLSLSLPGVDST 680
>TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose-responsive
element binding factor, partial (28%)
Length = 346
Score = 214 bits (544), Expect = 1e-56
Identities = 99/101 (98%), Positives = 99/101 (98%)
Frame = +1
Query: 1 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 60
MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF
Sbjct: 43 MDRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPF 222
Query: 61 TPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNST 101
TPEEDS IIRAHAR GNKWATIARILNGRTDNAVKNHWNST
Sbjct: 223 TPEEDSTIIRAHARCGNKWATIARILNGRTDNAVKNHWNST 345
>BF177776
Length = 384
Score = 200 bits (508), Expect = 2e-52
Identities = 102/105 (97%), Positives = 103/105 (97%)
Frame = +2
Query: 137 SAMSDPGPPAAISTDAPIAVPAAVVVPATDPATSLSLSLPGFDSSGSGSGSKQLFGAEFL 196
SA DPGPPAAISTDAPIAVPAAVVVPATDPATSLSLSLPGFDSSGSGSGSKQLFGAEFL
Sbjct: 5 SARGDPGPPAAISTDAPIAVPAAVVVPATDPATSLSLSLPGFDSSGSGSGSKQLFGAEFL 184
Query: 197 AVMQEMIRKEVRSYMSGMEEKNGACMPAEAIGNAVMKRMGISNVK 241
AVMQEMIRKEVRSYMSGMEEKNGAC+PAEAIGNAVMKRMGISNVK
Sbjct: 185 AVMQEMIRKEVRSYMSGMEEKNGACIPAEAIGNAVMKRMGISNVK 319
>TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose-responsive
element binding factor, partial (50%)
Length = 487
Score = 175 bits (443), Expect = 7e-45
Identities = 78/104 (75%), Positives = 89/104 (85%)
Frame = +2
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVEHRPFT 61
+R+KGPWS EED LTRLV+ HG +NW+ IS+ I GRSGKSCRLRWCNQLSP VEHRPF+
Sbjct: 161 ERIKGPWSAEEDRILTRLVERHGARNWSLISRYIKGRSGKSCRLRWCNQLSPTVEHRPFS 340
Query: 62 PEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
+ED II AHARFGN+WA IAR+L GRTDNAVKNHWNSTLKR+
Sbjct: 341 VQEDETIIAAHARFGNRWAAIARLLPGRTDNAVKNHWNSTLKRR 472
>NP675423 transcription factor MYB101 [Lotus japonicus]
Length = 1047
Score = 116 bits (290), Expect = 4e-27
Identities = 58/115 (50%), Positives = 71/115 (61%), Gaps = 2/115 (1%)
Frame = +1
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTPE 63
KGPW+PEED L +Q HG +W A+ K R GKSCRLRW N L P+++ FT E
Sbjct: 40 KGPWTPEEDAILVDYIQKHGHGSWRALPKLAGLNRCGKSCRLRWTNYLRPDIKRGKFTEE 219
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVD-TAVDHRP 117
E+ II HA GNKW+ IA L GRTDN +KN WN+ LK+K + V HRP
Sbjct: 220 EEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKKLLQMGLDPVTHRP 384
>NP675421 transcription factor MYB103 [Lotus japonicus]
Length = 924
Score = 115 bits (289), Expect = 5e-27
Identities = 55/111 (49%), Positives = 70/111 (62%), Gaps = 1/111 (0%)
Frame = +1
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTPE 63
KGPWS EED+ L +Q HG +W A+ + R GKSCRLRW N L P+++ F+ E
Sbjct: 40 KGPWSQEEDKVLVDYIQKHGHGSWRALPQLAGLNRCGKSCRLRWTNYLRPDIKRGKFSDE 219
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVD 114
E+ II HA GNKWATIA L GRTDN +KN WN+ LK+K + +D
Sbjct: 220 EEQLIINLHASLGNKWATIASHLPGRTDNEIKNLWNTHLKKKLKLLQMGLD 372
>TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial (41%)
Length = 1046
Score = 114 bits (285), Expect = 1e-26
Identities = 75/218 (34%), Positives = 105/218 (47%), Gaps = 5/218 (2%)
Frame = +2
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTPE 63
KG W+ EED+ L V +G NW + K R GKSCRLRW N L P ++ FT E
Sbjct: 131 KGTWTAEEDKKLIAYVTRYGCWNWRQLPKFAGLARCGKSCRLRWLNYLRPNIKRGNFTQE 310
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTAVDHRPLKRSAS 123
E+ IIR H + GN+W+TIA L GRTD+ VKNHW++TLKR +T V + K S S
Sbjct: 311 EEEIIIRMHKKLGNRWSTIAAELPGRTDSEVKNHWHTTLKRTVHEQNT-VTNEETKVSNS 487
Query: 124 VGAGCLNPSSPTGSAMSDPGPPAAISTD----APIAVPAAVVVPATDPATSLSLSLPGFD 179
P S S + P P +IS +P + + +D TS S + F+
Sbjct: 488 KNERDPVPKSGIASLLVTPPPATSISGSIGALSPFSTSSEFSCTTSDLTTSSSSEMLVFE 667
Query: 180 SSGSGSGSKQLFGAEFLAVMQEMIRKEVRSYMSGMEEK 217
F +F + E ++ Y++ + K
Sbjct: 668 DDFD-------FLDDFTENVNENFWSDLEPYLANISNK 760
>AV766688
Length = 608
Score = 108 bits (271), Expect = 6e-25
Identities = 59/140 (42%), Positives = 79/140 (56%), Gaps = 3/140 (2%)
Frame = +2
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTPE 63
KG W+ EED L V +G NW + K R GKSCRLRW N L P+++ FT E
Sbjct: 116 KGTWTAEEDRKLIAYVTRYGCWNWRQLPKFAGLARCGKSCRLRWLNYLRPDIKRGHFTQE 295
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKCSAVDTA--VDHRPLKRS 121
E+ IIR H GN+W+ IA L GRTDN VKNHW+++LKR+ + A + + +
Sbjct: 296 EEEIIIRMHKNLGNRWSIIAAELPGRTDNEVKNHWHTSLKRRVQQNEEAKVSNSKNEGPT 475
Query: 122 ASVGAGCLNPSSPTGSAMSD 141
+ G G L + P S +SD
Sbjct: 476 SGNGIGSLQVTPPATSQISD 535
>AV408163
Length = 425
Score = 106 bits (264), Expect = 4e-24
Identities = 49/102 (48%), Positives = 67/102 (65%), Gaps = 1/102 (0%)
Frame = +2
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
KG W+ EED+ L ++ HG W ++ K+ R GKSCRLRW N L P+++ FT E
Sbjct: 116 KGAWTKEEDDRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRPDLKRGNFTEE 295
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
ED II+ H+ GNKW+ IA L GRTDN +KN+WN+ ++RK
Sbjct: 296 EDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRK 421
>CN824947
Length = 738
Score = 105 bits (263), Expect = 5e-24
Identities = 53/123 (43%), Positives = 75/123 (60%), Gaps = 7/123 (5%)
Frame = +1
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
+G W+ EEDE LT+ +Q G +W ++ K+ R GKSCRLRW N L +++ + E
Sbjct: 190 RGRWTAEEDELLTKYIQASGEGSWRSLPKNAGLLRCGKSCRLRWINYLRGDLKRGNISDE 369
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKC------SAVDTAVDHRP 117
E+S I++ HA FGN+W+ IA + GRTDN +KN+WNS L RK S + VD P
Sbjct: 370 EESLIVKLHASFGNRWSLIASHMPGRTDNEIKNYWNSHLSRKLVYTFRGSTTKSIVDTPP 549
Query: 118 LKR 120
+R
Sbjct: 550 KRR 558
>TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete
Length = 1281
Score = 69.7 bits (169), Expect(2) = 2e-23
Identities = 31/61 (50%), Positives = 40/61 (64%), Gaps = 1/61 (1%)
Frame = +1
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTPE 63
KGPW+PEED+ L +Q HG +W A+ K R GKSCRLRW N L P+++ F+PE
Sbjct: 106 KGPWTPEEDQKLVEHIQKHGHGSWRALPKLAGLNRCGKSCRLRWTNYLRPDIKRGKFSPE 285
Query: 64 E 64
E
Sbjct: 286 E 288
Score = 55.1 bits (131), Expect(2) = 2e-23
Identities = 23/46 (50%), Positives = 32/46 (69%)
Frame = +2
Query: 60 FTPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
F +++ I+ H+ GNKW+TIA L GRTDN +KN WN+ LK+K
Sbjct: 275 FLQKKEQTILHLHSILGNKWSTIATHLPGRTDNEIKNFWNTHLKKK 412
>AV421932
Length = 402
Score = 102 bits (255), Expect = 5e-23
Identities = 49/95 (51%), Positives = 60/95 (62%), Gaps = 1/95 (1%)
Frame = +3
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAI-SKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPE 63
+G WSPEEDE L R + HG W+ + K+ R GKSCRLRW N L P++ FTPE
Sbjct: 117 RGLWSPEEDEKLIRYITTHGYGCWSEVPEKAGLQRCGKSCRLRWINYLRPDIRRGRFTPE 296
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHW 98
E+ II H GN+WA IA L GRTDN +KN+W
Sbjct: 297 EEKLIISLHGVVGNRWAHIASHLPGRTDNEIKNYW 401
>TC20215 similar to AAS10104 (AAS10104) MYB transcription factor, partial
(35%)
Length = 464
Score = 102 bits (254), Expect = 6e-23
Identities = 48/102 (47%), Positives = 67/102 (65%), Gaps = 1/102 (0%)
Frame = +2
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
+G WS EED+ LT ++ +G +W ++ K+ R GKSCRLRW N L +V+ T E
Sbjct: 68 RGRWSEEEDKILTDFIKQNGEGSWKSLPKNAGLLRCGKSCRLRWINYLREDVKRGNITSE 247
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRK 105
E+ I++ HA GN+W+ IA L GRTDN +KN+WNS L+RK
Sbjct: 248 EEEIIVKLHAALGNRWSVIAGHLPGRTDNEIKNYWNSHLRRK 373
>TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%)
Length = 532
Score = 102 bits (253), Expect = 8e-23
Identities = 47/101 (46%), Positives = 64/101 (62%), Gaps = 1/101 (0%)
Frame = +3
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTPE 63
KGPW+ EED L + HG W ++KS R+GKSCRLRW N L P+V TPE
Sbjct: 111 KGPWTIEEDLILINYIANHGEGVWNTLAKSAGLKRTGKSCRLRWLNYLRPDVRRGNITPE 290
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKR 104
E I+ HA++GN+W+ IA+ L GRTDN +KN W + +++
Sbjct: 291 EQLLIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQK 413
>AV413379
Length = 416
Score = 94.7 bits (234), Expect = 1e-20
Identities = 45/92 (48%), Positives = 59/92 (63%), Gaps = 1/92 (1%)
Frame = +3
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
KG W+ EED+ L ++ HG W ++ KS R GKSCRLRW N L P+++ FTP+
Sbjct: 141 KGAWTKEEDDRLIAYIRAHGEGCWRSLPKSAGLLRCGKSCRLRWINYLRPDLKRGNFTPQ 320
Query: 64 EDSAIIRAHARFGNKWATIARILNGRTDNAVK 95
ED II+ H+ GNKW+ IA L GRTDN +K
Sbjct: 321 EDELIIKLHSLLGNKWSLIAGRLAGRTDNEIK 416
>AV417320
Length = 369
Score = 84.3 bits (207), Expect = 2e-17
Identities = 39/80 (48%), Positives = 50/80 (61%), Gaps = 1/80 (1%)
Frame = +3
Query: 5 KGPWSPEEDEALTRLVQVHGPKNWTAISKSIPG-RSGKSCRLRWCNQLSPEVEHRPFTPE 63
KGPW+ EED+ L +Q HG NW A+ K R GKSCRLRW N L P+++ FT E
Sbjct: 117 KGPWTSEEDQILISYIQKHGHGNWRALPKHAGLLRCGKSCRLRWINYLRPDIKRGNFTAE 296
Query: 64 EDSAIIRAHARFGNKWATIA 83
E+ II+ H GN+W+ IA
Sbjct: 297 EEELIIKMHELLGNRWSAIA 356
>CB828471
Length = 525
Score = 79.3 bits (194), Expect = 5e-16
Identities = 43/128 (33%), Positives = 67/128 (51%), Gaps = 10/128 (7%)
Frame = +1
Query: 38 RSGKSCRLRWCNQLSPEVEHRPFTPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNH 97
R GKSCRLRW N L P+++ F+ EE+ II H GN+W+ IA L GRTDN +KN
Sbjct: 22 RCGKSCRLRWANYLRPDIKRGNFSREEEDEIINLHELLGNRWSAIAARLPGRTDNEIKNV 201
Query: 98 WNSTLKRKCSA----------VDTAVDHRPLKRSASVGAGCLNPSSPTGSAMSDPGPPAA 147
W++ LK++ +D + ++ K+ G P SP + S+ ++
Sbjct: 202 WHTHLKKRLQPQTKQKQTNLDLDASKSNKDAKKEHQEDEG---PRSPQQCSSSNSDNMSS 372
Query: 148 ISTDAPIA 155
++ + IA
Sbjct: 373 LTNSSSIA 396
Score = 30.4 bits (67), Expect = 0.28
Identities = 15/55 (27%), Positives = 30/55 (54%)
Frame = +1
Query: 2 DRVKGPWSPEEDEALTRLVQVHGPKNWTAISKSIPGRSGKSCRLRWCNQLSPEVE 56
D +G +S EE++ + L ++ G + W+AI+ +PGR+ + W L ++
Sbjct: 70 DIKRGNFSREEEDEIINLHELLGNR-WSAIAARLPGRTDNEIKNVWHTHLKKRLQ 231
>BI420780
Length = 475
Score = 68.2 bits (165), Expect = 1e-12
Identities = 35/86 (40%), Positives = 45/86 (51%), Gaps = 3/86 (3%)
Frame = +1
Query: 8 WSPEEDEALTRLVQVHGPKNWTAISKSIP---GRSGKSCRLRWCNQLSPEVEHRPFTPEE 64
WS EED L VQ +GP+ W +S+ + R KSC RW N L P ++ T EE
Sbjct: 211 WSSEEDALLHAYVQQYGPREWNLVSQRMNTPLNRDTKSCLERWKNYLKPGIKKGSLTKEE 390
Query: 65 DSAIIRAHARFGNKWATIARILNGRT 90
+I A +GNKW IA + GRT
Sbjct: 391 QRLVILLQANYGNKWKKIAAEVPGRT 468
>AV776018
Length = 476
Score = 55.5 bits (132), Expect = 8e-09
Identities = 30/61 (49%), Positives = 37/61 (60%), Gaps = 2/61 (3%)
Frame = +1
Query: 60 FTPEEDSAIIRAHARFGNKWATIARILNGRTDNAVKNHWNSTLKRKC--SAVDTAVDHRP 117
F EED+ II HA GN+W+ A GRTDN +KN WNS LK+K +D V H+P
Sbjct: 13 FHKEEDNLIIELHAVLGNRWSQDATQWPGRTDNEIKNLWNSCLKKKLRQRGID-PVTHKP 189
Query: 118 L 118
L
Sbjct: 190 L 192
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.129 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,986,883
Number of Sequences: 28460
Number of extensions: 57662
Number of successful extensions: 488
Number of sequences better than 10.0: 107
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 472
length of query: 241
length of database: 4,897,600
effective HSP length: 88
effective length of query: 153
effective length of database: 2,393,120
effective search space: 366147360
effective search space used: 366147360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0321.10


