

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0320b.5
(930 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
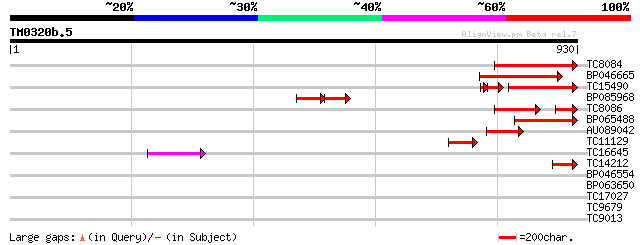
Score E
Sequences producing significant alignments: (bits) Value
TC8084 similar to UP|PIF1_YEAST (P07271) DNA repair and recombin... 184 6e-47
BP046665 174 8e-44
TC15490 weakly similar to UP|Q9LTU4 (Q9LTU4) Helicase-like prote... 146 3e-41
BP085968 82 3e-31
TC8086 similar to UP|DMC1_HUMAN (Q14565) Meiotic recombination p... 95 2e-28
BP065488 97 2e-20
AU089042 71 9e-13
TC11129 weakly similar to UP|PDA6_MEDSA (P38661) Probable protei... 65 5e-11
TC16645 54 1e-07
TC14212 similar to UP|Q9FN61 (Q9FN61) Gb|AAF07369.1, partial (10%) 47 1e-05
BP046554 38 0.006
BP063650 31 0.22
TC17027 similar to UP|Q93XA5 (Q93XA5) Homeodomain leucine zipper... 28 6.7
TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, p... 28 6.7
TC9013 28 6.7
>TC8084 similar to UP|PIF1_YEAST (P07271) DNA repair and recombination
protein PIF1, mitochondrial precursor, partial (4%)
Length = 560
Score = 184 bits (467), Expect = 6e-47
Identities = 89/135 (65%), Positives = 109/135 (79%)
Frame = +1
Query: 796 IDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQF 855
I + LCNGTRLIV L Y+I AT ++G+ G ++IPRL + PSD+G PFKF RR F
Sbjct: 43 IQKFENLCNGTRLIVVDLGTYVIKATVITGTNIGDDIFIPRLDMVPSDSGYPFKFERR*F 222
Query: 856 PITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVS 915
PI++CFAMTINKSQGQSLSHV LYL RPVFTHGQLYVALSRV+SRK LK+L++D++ V+
Sbjct: 223 PISLCFAMTINKSQGQSLSHVSLYLSRPVFTHGQLYVALSRVRSRKGLKLLVLDEEEKVT 402
Query: 916 NCTRNVVYEEVFQNI 930
N T+NVVY EVF+NI
Sbjct: 403 NTTKNVVYREVFENI 447
>BP046665
Length = 524
Score = 174 bits (440), Expect = 8e-44
Identities = 85/136 (62%), Positives = 105/136 (76%)
Frame = -1
Query: 771 DLKCSGIPNHRIVLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGK 830
D+ C GIPNH+I LK G PIML++NI Q+ G CNGTRLIV+ L +I AT ++ + G
Sbjct: 524 DITCFGIPNHKITLKEGAPIMLLRNIYQAVGFCNGTRLIVADLGTNVIKATVIT*TNIGD 345
Query: 831 PVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVGLYLPRPVFTHGQL 890
++IPR+ + PSD+G PFKF RRQFPI++C AMTINKSQGQSLSHVGLYL R VFTHGQL
Sbjct: 344 DIFIPRMDMVPSDSGYPFKFERRQFPISLCSAMTINKSQGQSLSHVGLYLSRHVFTHGQL 165
Query: 891 YVALSRVKSRKRLKIL 906
YVAL K +KR +I+
Sbjct: 164 YVAL-HAKIKKRTQII 120
Score = 38.9 bits (89), Expect = 0.004
Identities = 18/45 (40%), Positives = 31/45 (68%)
Frame = -2
Query: 879 YLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVY 923
Y+ +++H LS ++SRK LK+L++D++ V+N T+NVVY
Sbjct: 202 YIFLAMYSHMVSCTLLSMLRSRKGLKLLVLDEEEKVTNTTKNVVY 68
>TC15490 weakly similar to UP|Q9LTU4 (Q9LTU4) Helicase-like protein, partial
(8%)
Length = 634
Score = 146 bits (368), Expect(3) = 3e-41
Identities = 69/113 (61%), Positives = 90/113 (79%)
Frame = +2
Query: 818 IVATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQSLSHVG 877
I T ++G+ G + IPR+ + PSD+ PFKF RRQ PI++CFAMTINKSQG+SLSHVG
Sbjct: 137 IQVTVITGTHIGDDISIPRMDMVPSDSSYPFKFERRQSPISLCFAMTINKSQGRSLSHVG 316
Query: 878 LYLPRPVFTHGQLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
LYL RPV THG LYVAL RV+SRK LK+L++D++ ++N T+NVVY E+F+NI
Sbjct: 317 LYLSRPVSTHG*LYVALPRVRSRK*LKLLVLDEEEKMTNTTKNVVYREIFENI 475
Score = 38.9 bits (89), Expect(3) = 3e-41
Identities = 18/26 (69%), Positives = 22/26 (84%)
Frame = +1
Query: 785 KVGVPIMLIQNIDQSAGLCNGTRLIV 810
K G IML++NI Q++GLCNGTRLIV
Sbjct: 43 KEGALIMLLRNIVQASGLCNGTRLIV 120
Score = 22.3 bits (46), Expect(3) = 3e-41
Identities = 8/14 (57%), Positives = 10/14 (71%)
Frame = +3
Query: 772 LKCSGIPNHRIVLK 785
+ CS IPNH+I K
Sbjct: 3 ITCSDIPNHKITEK 44
>BP085968
Length = 341
Score = 81.6 bits (200), Expect(2) = 3e-31
Identities = 38/47 (80%), Positives = 43/47 (90%)
Frame = +2
Query: 471 FFFLYGFGGSGKTFVWNTLSAALRSEGKIVLNVASSGIASLLLPGGR 517
F+FLYGFGGSGKTFVWNT S+ LRS+G +VLNVASSGIAS LLPGG+
Sbjct: 74 FYFLYGFGGSGKTFVWNTWSSGLRSQGLMVLNVASSGIASWLLPGGK 214
Score = 71.6 bits (174), Expect(2) = 3e-31
Identities = 33/43 (76%), Positives = 39/43 (89%)
Frame = +3
Query: 517 RTAHSRFSIPITIHESSTCNVRQGSHKAEMLQKASLIIWDEAP 559
RTAHSRFSI I+I++ STCN++QGS KAE+LQKAS IIWDEAP
Sbjct: 213 RTAHSRFSIHISINDISTCNIKQGSQKAELLQKASSIIWDEAP 341
>TC8086 similar to UP|DMC1_HUMAN (Q14565) Meiotic recombination protein
DMC1/LIM15 homolog, partial (12%)
Length = 628
Score = 94.7 bits (234), Expect(2) = 2e-28
Identities = 44/75 (58%), Positives = 56/75 (74%)
Frame = +1
Query: 796 IDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTPSDTGLPFKFSRRQF 855
I + LC+GTRLIV L Y+I AT ++G+ G ++IPRL + PSD+G PFKF RR F
Sbjct: 151 IQKFENLCHGTRLIVVDLGTYVIKATVITGTNIGDDIFIPRLDMVPSDSGYPFKFERR*F 330
Query: 856 PITVCFAMTINKSQG 870
PI++CFAMTINKSQG
Sbjct: 331 PISLCFAMTINKSQG 375
Score = 49.3 bits (116), Expect(2) = 2e-28
Identities = 22/36 (61%), Positives = 31/36 (86%)
Frame = +3
Query: 895 SRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
SRV+SRK LK+L++D++ V+N T+NVVY EVF+NI
Sbjct: 369 SRVRSRKGLKLLVLDEEEKVTNTTKNVVYREVFENI 476
>BP065488
Length = 439
Score = 96.7 bits (239), Expect = 2e-20
Identities = 52/104 (50%), Positives = 73/104 (70%), Gaps = 2/104 (1%)
Frame = -1
Query: 829 GKPVYIPRLSLTPSDTGLPFKFSRRQFPITVCFAMTINKSQGQ-SLSHVGLYLPRPVFTH 887
G ++IPR+++ PS +G P KF R QFPI++CFAMTINKSQ V L ++H
Sbjct: 379 GDDIFIPRMNMVPSVSGDPLKFERCQFPISLCFAMTINKSQXSVHYPTVALISFLACYSH 200
Query: 888 -GQLYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
+LYVALS V+SRK LK+L++ ++ V+N T+N VY+EVF+NI
Sbjct: 199 MDKLYVALSGVRSRKGLKLLVLGEEEKVTNATKNEVYQEVFKNI 68
>AU089042
Length = 191
Score = 70.9 bits (172), Expect = 9e-13
Identities = 36/61 (59%), Positives = 43/61 (70%)
Frame = +3
Query: 783 VLKVGVPIMLIQNIDQSAGLCNGTRLIVSALTPYIIVATALSGSKTGKPVYIPRLSLTPS 842
VLK GVP+ML+ N+ S GLCNGTRLIV L P +I AT LSG+ G VYI ++L PS
Sbjct: 6 VLKXGVPVMLMXNLXISTGLCNGTRLIVDYLGPNVIGATILSGTHIGXVVYISMMNLXPS 185
Query: 843 D 843
D
Sbjct: 186 D 188
>TC11129 weakly similar to UP|PDA6_MEDSA (P38661) Probable protein disulfide
isomerase A6 precursor (P5) , partial (7%)
Length = 600
Score = 65.1 bits (157), Expect = 5e-11
Identities = 27/46 (58%), Positives = 37/46 (79%)
Frame = +2
Query: 721 PTLESVEEVNNFMMSMIPGEETKYLSYDTPCRSDEDSEIDAEWFTS 766
PTLESVE+VN FM+ ++PG T+YLS DT C+ DED+E+ + WFT+
Sbjct: 458 PTLESVEKVNEFMLDLLPGNTTEYLSSDTTCKYDEDTELQS*WFTT 595
>TC16645
Length = 596
Score = 53.5 bits (127), Expect = 1e-07
Identities = 36/95 (37%), Positives = 47/95 (48%)
Frame = +2
Query: 227 TRREEV*RMLNIRLCLFIRKISKYGSQEREVSPLVVFNISL*AWARYIT*GFY*PNREAV 286
TRR E+ M N L F G ++ LV ++ L W R FY*P ++ V
Sbjct: 26 TRRGEISHMQNFLLSFFTTNTQPNGFLSKKDFLLVGCHLLLQEWERITICVFY*PCKKVV 205
Query: 287 TVLQV*ELSKGLFIQHSKMRAMLWD*WKMRGSMLM 321
TVL+ *+ + FI S M A WD W+M SMLM
Sbjct: 206 TVLKA*KRLRVWFIPLSMMHAKQWDCWRMIASMLM 310
>TC14212 similar to UP|Q9FN61 (Q9FN61) Gb|AAF07369.1, partial (10%)
Length = 613
Score = 47.4 bits (111), Expect = 1e-05
Identities = 24/41 (58%), Positives = 29/41 (70%)
Frame = +2
Query: 890 LYVALSRVKSRKRLKILIVDDKGVVSNCTRNVVYEEVFQNI 930
LYVA+SRVKS+ LKILI D T+N+VY+EVFQ I
Sbjct: 2 LYVAVSRVKSKDGLKILISSDGTSTPGATKNIVYKEVFQKI 124
>BP046554
Length = 556
Score = 38.1 bits (87), Expect = 0.006
Identities = 26/45 (57%), Positives = 29/45 (63%)
Frame = -1
Query: 48 KFKGMSVLGI*ELKIDLIFVLEFSR*SSII*CLI*GKARYLVLLM 92
KFKG+ LGI* K FV FS+*SSI LI* K +Y VLLM
Sbjct: 205 KFKGLCNLGI*M*KTVQTFVFVFSK*SSIDSYLI*KKGKYSVLLM 71
>BP063650
Length = 497
Score = 30.8 bits (68), Expect(2) = 0.22
Identities = 13/31 (41%), Positives = 20/31 (63%)
Frame = +1
Query: 602 QILPVILKGSRSEIISSSANSSYLWKHCKVM 632
+ L V+ KG +II + N+SYL HC+V+
Sbjct: 58 KFLTVVYKGRMQDIIHAIVNASYL*DHCQVL 150
Score = 20.8 bits (42), Expect(2) = 0.22
Identities = 9/10 (90%), Positives = 9/10 (90%)
Frame = +3
Query: 594 VVLGGDFRQI 603
VVL GDFRQI
Sbjct: 33 VVLQGDFRQI 62
>TC17027 similar to UP|Q93XA5 (Q93XA5) Homeodomain leucine zipper protein
HDZ1 (Fragment), partial (34%)
Length = 854
Score = 28.1 bits (61), Expect = 6.7
Identities = 15/37 (40%), Positives = 19/37 (50%)
Frame = +1
Query: 737 IPGEETKYLSYDTPCRSDEDSEIDAEWFTSEFLNDLK 773
IPG ++K LSYD C D +D S F +LK
Sbjct: 4 IPGSDSKELSYDC-CFKSSDDGVDGTTTASLFAENLK 111
>TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partial
(54%)
Length = 710
Score = 28.1 bits (61), Expect = 6.7
Identities = 11/36 (30%), Positives = 21/36 (57%)
Frame = -3
Query: 166 WLMASVLNTFPKNIRTALPLMMMVQMLEMENRTKMK 201
W ++ T + +RTA+P+MM + ++ R KM+
Sbjct: 276 WNFTTLGRTMSQKMRTAMPMMMRAVKMSLKMRLKMQ 169
>TC9013
Length = 766
Score = 28.1 bits (61), Expect = 6.7
Identities = 14/26 (53%), Positives = 16/26 (60%)
Frame = +3
Query: 905 ILIVDDKGVVSNCTRNVVYEEVFQNI 930
ILI D SN T NVVY EVF+ +
Sbjct: 627 ILICDGDDSNSNSTSNVVYTEVFRTV 704
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.344 0.151 0.484
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,281,243
Number of Sequences: 28460
Number of extensions: 203277
Number of successful extensions: 1734
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 1717
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1733
length of query: 930
length of database: 4,897,600
effective HSP length: 99
effective length of query: 831
effective length of database: 2,080,060
effective search space: 1728529860
effective search space used: 1728529860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0320b.5


