

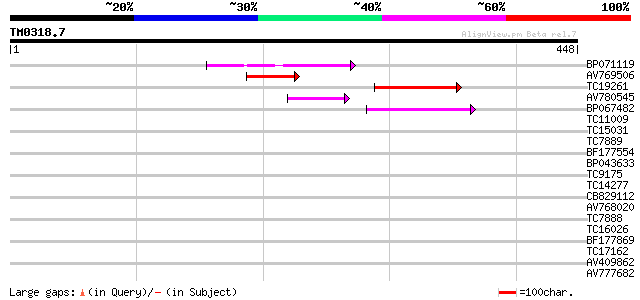
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.7
(448 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP071119 87 4e-18
AV769506 86 1e-17
TC19261 similar to UP|BAD06577 (BAD06577) Cell wall protein Awa1... 85 2e-17
AV780545 44 5e-05
BP067482 42 2e-04
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 38 0.004
TC15031 weakly similar to UP|Q69107 (Q69107) LAT protein, partia... 38 0.004
TC7889 similar to UP|Q40336 (Q40336) Proline-rich cell wall prot... 38 0.004
BF177554 37 0.005
BP043633 36 0.011
TC9175 weakly similar to UP|INAD_DROME (Q24008) Inactivation-no-... 35 0.025
TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_1... 35 0.033
CB829112 35 0.033
AV768020 34 0.056
TC7888 similar to UP|Q40336 (Q40336) Proline-rich cell wall prot... 33 0.073
TC16026 similar to UP|Q9SGZ3 (Q9SGZ3) F28K19.29, partial (32%) 33 0.073
BF177869 33 0.096
TC17162 similar to UP|Q8H5F1 (Q8H5F1) OJ1165_F02.16 protein, par... 32 0.16
AV409862 32 0.16
AV777682 32 0.16
>BP071119
Length = 412
Score = 87.4 bits (215), Expect = 4e-18
Identities = 49/118 (41%), Positives = 65/118 (54%)
Frame = +2
Query: 156 FPGGPVELSLLTYYADHKAPWAWHALLRTDERYVDRRQLKVATAGGKVWNLAYDGDSDSH 215
+ GGP +L LLT Y +H A W + R + Y R LK+ G + +
Sbjct: 80 YEGGPFDLCLLTKYGEHIARDIWRSYKRPN--YETRGVLKIYNHGRMCHPVVHHA----- 238
Query: 216 RRVRELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITLDDVSA 273
R +R + GL + C+ P D +I A VERWH ETSSFH+P+GEMTITL+DVSA
Sbjct: 239 RYIRGAVHNAGLRWVMKCTSPSVDQSIISAFVERWHPETSSFHLPWGEMTITLEDVSA 412
>AV769506
Length = 345
Score = 86.3 bits (212), Expect = 1e-17
Identities = 41/42 (97%), Positives = 41/42 (97%)
Frame = -3
Query: 188 YVDRRQLKVATAGGKVWNLAYDGDSDSHRRVRELIEQTGLHQ 229
YVDRRQLKVATAGGKVWNLA DGDSDSHRRVRELIEQTGLHQ
Sbjct: 343 YVDRRQLKVATAGGKVWNLACDGDSDSHRRVRELIEQTGLHQ 218
>TC19261 similar to UP|BAD06577 (BAD06577) Cell wall protein Awa1p, partial
(3%)
Length = 495
Score = 85.1 bits (209), Expect = 2e-17
Identities = 44/69 (63%), Positives = 51/69 (73%)
Frame = -1
Query: 289 GERDECAALCAQLMGGSVGIYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYEDAARIWLV 348
GERDE AALCAQL+GGSV Y AEF++ QT+RF L+T +A+ RYEDAARIWLV
Sbjct: 495 GERDEVAALCAQLLGGSVAAYLAEFESAGGQTLRFLTLKTMSTSAMDGGRYEDAARIWLV 316
Query: 349 NQLGATLFA 357
N L ATL A
Sbjct: 315 NPLAATLVA 289
>AV780545
Length = 555
Score = 43.9 bits (102), Expect = 5e-05
Identities = 23/49 (46%), Positives = 29/49 (58%)
Frame = +1
Query: 220 ELIEQTGLHQLPYCSYPVTDAGLILALVERWHEETSSFHMPFGEMTITL 268
E +E+ G L D LI ALVERW ET++FH+ GEMT+TL
Sbjct: 409 E*VERAGFGYLRSIPAISLDNPLISALVERWRRETNTFHLNVGEMTVTL 555
>BP067482
Length = 291
Score = 42.0 bits (97), Expect = 2e-04
Identities = 28/87 (32%), Positives = 41/87 (46%), Gaps = 1/87 (1%)
Frame = +2
Query: 283 FYTPGRGERDECAALCAQLMGGSVG-IYEAEFDTNRSQTIRFGVLQTRYEAALAEHRYED 341
F++ G R+E A AQL+ + I EFD R ++RF LQ E + +
Sbjct: 23 FFSFGNPTREEAAPAVAQLLDVDIETI**EEFDACRGPSLRFSFLQAIVETHVEANNEVP 202
Query: 342 AARIWLVNQLGATLFASKSGGYHTTVY 368
A R +++ +G TL KS Y VY
Sbjct: 203 ACRAYMLRWIGMTLVCDKSNTYIGAVY 283
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 37.7 bits (86), Expect = 0.004
Identities = 33/109 (30%), Positives = 42/109 (38%)
Frame = -2
Query: 5 KRSHASSEDVGATEDRHRRLHASSRRGDHAATSQAVEASAPVVSPPSPMIEVPLVDYPPS 64
K ASS AT R R S +++S + +S+ SPP P PPS
Sbjct: 300 KSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPR--------PPS 145
Query: 65 PTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASS 113
+PPS SS SS + SS SS SS +SS
Sbjct: 144 ---------TPPSSSSSSSSSSSSPPPGASSSSSSSPSSSSPSSSSSSS 25
Score = 34.3 bits (77), Expect = 0.043
Identities = 21/77 (27%), Positives = 34/77 (43%), Gaps = 5/77 (6%)
Frame = +2
Query: 81 ESSGEESSGEESSGEESSG-----EESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
+ GE+ G+E +++ G ++ EE G G GG + P DDD
Sbjct: 38 DDDGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGD---PDDDDDDDDDDD 208
Query: 136 EQGEQGAQGGEEDLIQR 152
E+ E+ G E L++R
Sbjct: 209 EEEEEEEDLGTEYLVRR 259
Score = 31.2 bits (69), Expect = 0.36
Identities = 18/49 (36%), Positives = 18/49 (36%)
Frame = -2
Query: 78 PMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPP 126
P SS SS SS SSG S SS S S PPP
Sbjct: 237 PKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSPPP 91
Score = 30.8 bits (68), Expect = 0.47
Identities = 20/67 (29%), Positives = 33/67 (48%)
Frame = +2
Query: 82 SSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQGEQG 141
+S SG+E ++ GE+ G+E + + G GG D+D D++ E G +G
Sbjct: 2 NSRTRMSGDED--DDDDGEDDDGDEDDDDDAPG-GGDDDD-------DEEDEEEEGGVEG 151
Query: 142 AQGGEED 148
+GG D
Sbjct: 152 GRGGGGD 172
>TC15031 weakly similar to UP|Q69107 (Q69107) LAT protein, partial (27%)
Length = 997
Score = 37.7 bits (86), Expect = 0.004
Identities = 23/62 (37%), Positives = 26/62 (41%)
Frame = +3
Query: 64 SPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSI 123
SP+V T VS P+P S +SG S SS SSG SS G S I
Sbjct: 525 SPSVSSSTFVSNPNPSPISKASTASGPSVSSSSSSSSSGPSPNSSGYPSSAAGTSPPSPI 704
Query: 124 PP 125
P
Sbjct: 705 KP 710
>TC7889 similar to UP|Q40336 (Q40336) Proline-rich cell wall protein,
partial (66%)
Length = 1158
Score = 37.7 bits (86), Expect = 0.004
Identities = 20/43 (46%), Positives = 27/43 (62%)
Frame = +1
Query: 40 VEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVES 82
V S P+V+PP+P P + PP+PT P VV+PPSP E+
Sbjct: 370 VVPSPPIVTPPTP---TPPIVTPPTPT---PPVVTPPSPPTET 480
Score = 33.1 bits (74), Expect = 0.096
Identities = 18/38 (47%), Positives = 22/38 (57%)
Frame = +1
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVES 82
P+V+PP+P P V PPSP E P PP P+V S
Sbjct: 415 PIVTPPTP---TPPVVTPPSPPTETP--CPPPPPVVPS 513
Score = 32.3 bits (72), Expect = 0.16
Identities = 16/35 (45%), Positives = 18/35 (50%)
Frame = +1
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPM 79
PVV+PPSP E P PP P V P P P+
Sbjct: 445 PVVTPPSPPTETPCP--PPPPVVPSPPPAQPTCPI 543
Score = 31.2 bits (69), Expect = 0.36
Identities = 17/43 (39%), Positives = 23/43 (52%), Gaps = 9/43 (20%)
Frame = +1
Query: 45 PVVSPP----SPMIEVPLVDYPP-----SPTVEIPTVVSPPSP 78
P+V PP +P I P + YPP P V P +V+PP+P
Sbjct: 280 PIVYPPPYVPTPPIVKPPIVYPPPYVPLPPVVPSPPIVTPPTP 408
Score = 30.0 bits (66), Expect = 0.81
Identities = 14/36 (38%), Positives = 21/36 (57%), Gaps = 2/36 (5%)
Frame = +1
Query: 45 PVVSPPSPMIEVPLVDYPP--SPTVEIPTVVSPPSP 78
P+V PP + P+V PP +P P +V+PP+P
Sbjct: 331 PIVYPPPYVPLPPVVPSPPIVTPPTPTPPIVTPPTP 438
Score = 28.1 bits (61), Expect = 3.1
Identities = 14/32 (43%), Positives = 17/32 (52%)
Frame = +1
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPP 76
PVV P + P + YPP P V P +V PP
Sbjct: 190 PVVPVTPPYVPKPPIVYPP-PYVPKPPIVKPP 282
Score = 26.9 bits (58), Expect = 6.8
Identities = 15/37 (40%), Positives = 17/37 (45%), Gaps = 5/37 (13%)
Frame = +1
Query: 45 PVVSPP-----SPMIEVPLVDYPPSPTVEIPTVVSPP 76
PVV PP P++ P V PP V P V PP
Sbjct: 82 PVVKPPPYVPKPPVVSPPYVPKPPVVPVTPPYVPKPP 192
>BF177554
Length = 246
Score = 37.4 bits (85), Expect = 0.005
Identities = 30/94 (31%), Positives = 37/94 (38%)
Frame = +1
Query: 85 EESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPPEQGEQGAQG 144
+E S EE +GE S EE EE E G D D+ PE+ E
Sbjct: 25 DEDSCEEEAGESSEEEEEEEEEGEEEEEEDDGPGDRDA-----------QPEEEE----- 156
Query: 145 GEEDLIQRLPPFPGGPVELSLLTYYADHKAPWAW 178
EED GGP ++SLL + H A W
Sbjct: 157 -EEDF-----SLLGGPYDISLLKSFKGHVALRVW 240
>BP043633
Length = 542
Score = 36.2 bits (82), Expect = 0.011
Identities = 35/109 (32%), Positives = 43/109 (39%), Gaps = 7/109 (6%)
Frame = +3
Query: 23 RLHASSRRGDHAATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVES 82
RLHASS T+ + ASA PP P P SP V P V PP P +S
Sbjct: 228 RLHASS-------TTTSASASAA*SQPPPPFPPNTTATSPSSPVVLPPISVPPPIP--DS 380
Query: 83 SGEESSGEESSGEESSGE-------ESSGEESSGEASSGMGGSDEDSIP 124
+ G ++ SS E S SS +SS S S+P
Sbjct: 381 FPSTAVGATTTAASSSVEPLFLPPQAVSSSSSSSSSSSPASSSVPTSLP 527
>TC9175 weakly similar to UP|INAD_DROME (Q24008)
Inactivation-no-after-potential D protein, partial (5%)
Length = 692
Score = 35.0 bits (79), Expect = 0.025
Identities = 24/91 (26%), Positives = 39/91 (42%)
Frame = +2
Query: 58 LVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASSGMGG 117
LV+Y T + +V P P E E E+ + EE EE EE + E + G
Sbjct: 5 LVEYVYRRTKKQAKIVPQPEPEPEKKEEGKEEEKPAAEEGKPEEKKEEEKAAEEAKKEEG 184
Query: 118 SDEDSIPPPTVDDDVLPPEQGEQGAQGGEED 148
+E +D + GE+G + G+++
Sbjct: 185 GEE--------KNDNKEEKGGEEGKEEGKKE 253
>TC14277 similar to UP|Q9SHY3 (Q9SHY3) F1E22.9 (AT1G65720/F1E22_13), partial
(33%)
Length = 941
Score = 34.7 bits (78), Expect = 0.033
Identities = 19/75 (25%), Positives = 34/75 (45%)
Frame = +3
Query: 34 AATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESS 93
AA+ +S+ PP P PP+P+ +P SPPSP + S +S +
Sbjct: 84 AASRSPPSSSSSPSPPPPPHAPAAASSSPPTPSATLPPTPSPPSPRSDPSPLSTSTTNPT 263
Query: 94 GEESSGEESSGEESS 108
S+ +++ +S+
Sbjct: 264 TSSSTTPKTTTNKSN 308
>CB829112
Length = 234
Score = 34.7 bits (78), Expect = 0.033
Identities = 21/64 (32%), Positives = 32/64 (49%), Gaps = 2/64 (3%)
Frame = +3
Query: 35 ATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSP--PSPMVESSGEESSGEES 92
ATS A+ + AP+ PP P++ P + PP P T + P PS M + S + SS
Sbjct: 36 ATS*ALVSLAPLAPPPPPLLLPPPLLPPPPPPPPPATHILPMTPSRMTQMSSKRSSASSE 215
Query: 93 SGEE 96
+ +
Sbjct: 216AAPQ 227
>AV768020
Length = 268
Score = 33.9 bits (76), Expect = 0.056
Identities = 18/63 (28%), Positives = 27/63 (42%)
Frame = -2
Query: 33 HAATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEES 92
H +SQ+ PP P P + P PT++IP+ P ++ EE+ EE
Sbjct: 195 HRCSSQSTTTRWYCTVPPPPATHSPPLHLPSPPTLQIPSSTMQTEPTRKADKEETQREEI 16
Query: 93 SGE 95
E
Sbjct: 15 ERE 7
>TC7888 similar to UP|Q40336 (Q40336) Proline-rich cell wall protein,
partial (60%)
Length = 913
Score = 33.5 bits (75), Expect = 0.073
Identities = 18/38 (47%), Positives = 24/38 (62%)
Frame = +3
Query: 40 VEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPS 77
V S P+V+PP+P P + PP+PT P VV+PPS
Sbjct: 816 VVPSPPIVTPPTP---TPPIVTPPTPT---PPVVTPPS 911
Score = 31.2 bits (69), Expect = 0.36
Identities = 17/43 (39%), Positives = 23/43 (52%), Gaps = 9/43 (20%)
Frame = +3
Query: 45 PVVSPP----SPMIEVPLVDYPP-----SPTVEIPTVVSPPSP 78
P+V PP +P I P + YPP P V P +V+PP+P
Sbjct: 726 PIVYPPPYVPTPPIVKPPIVYPPPYVPLPPVVPSPPIVTPPTP 854
Score = 30.0 bits (66), Expect = 0.81
Identities = 14/36 (38%), Positives = 21/36 (57%), Gaps = 2/36 (5%)
Frame = +3
Query: 45 PVVSPPSPMIEVPLVDYPP--SPTVEIPTVVSPPSP 78
P+V PP + P+V PP +P P +V+PP+P
Sbjct: 777 PIVYPPPYVPLPPVVPSPPIVTPPTPTPPIVTPPTP 884
Score = 28.1 bits (61), Expect = 3.1
Identities = 16/34 (47%), Positives = 18/34 (52%), Gaps = 2/34 (5%)
Frame = +3
Query: 45 PVVSP--PSPMIEVPLVDYPPSPTVEIPTVVSPP 76
PV P P P I P + YPP P V P +V PP
Sbjct: 630 PVTPPYVPKPPIVKPPIVYPP-PYVPKPPIVKPP 728
Score = 26.9 bits (58), Expect = 6.8
Identities = 15/37 (40%), Positives = 17/37 (45%), Gaps = 5/37 (13%)
Frame = +3
Query: 45 PVVSPP-----SPMIEVPLVDYPPSPTVEIPTVVSPP 76
PVV PP P++ P V PP V P V PP
Sbjct: 513 PVVKPPPYVPKPPVVSPPYVPKPPVVPVTPPYVPKPP 623
>TC16026 similar to UP|Q9SGZ3 (Q9SGZ3) F28K19.29, partial (32%)
Length = 1183
Score = 33.5 bits (75), Expect = 0.073
Identities = 27/92 (29%), Positives = 38/92 (40%), Gaps = 11/92 (11%)
Frame = +1
Query: 42 ASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSG-------EESSG 94
+S+ + PP+P+ + PS + P P PM ES E S E+
Sbjct: 832 SSSSWLQPPNPISD-------PSDPISDPDPTEQPPPMAESESESPSQHHTQLP*SEADS 990
Query: 95 EESSGEESSGEE----SSGEASSGMGGSDEDS 122
+E+ GE S E S EA MG D+ S
Sbjct: 991 DEARGESSKTHEAIQRSEAEALGKMGR*DQAS 1086
>BF177869
Length = 509
Score = 33.1 bits (74), Expect = 0.096
Identities = 23/69 (33%), Positives = 34/69 (48%), Gaps = 1/69 (1%)
Frame = +2
Query: 55 EVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSGEESSGEASSG 114
E+P+ ++PP VEI E + ++G SS SS SSG SS ++ SG
Sbjct: 170 EMPMSEFPP---VEI-----------EKDKDVAAGHASSSSGSSSSSSSGSSSSSDSDSG 307
Query: 115 -MGGSDEDS 122
GSD ++
Sbjct: 308 SSSGSDSEA 334
>TC17162 similar to UP|Q8H5F1 (Q8H5F1) OJ1165_F02.16 protein, partial (40%)
Length = 784
Score = 32.3 bits (72), Expect = 0.16
Identities = 25/87 (28%), Positives = 33/87 (37%), Gaps = 3/87 (3%)
Frame = +1
Query: 31 GDHAATSQAVEASAPVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGE---ES 87
GD TS++ +++P S SP PP + PT SPP P G
Sbjct: 202 GDPTPTSRSTPSASPTPSAASPD--------PPRKSATPPTASSPPPPKAGRVGAARFSP 357
Query: 88 SGEESSGEESSGEESSGEESSGEASSG 114
+G S+ SS EE SG
Sbjct: 358 AGRSSASRRSSEPPPD*EERRSGGGSG 438
>AV409862
Length = 420
Score = 32.3 bits (72), Expect = 0.16
Identities = 23/83 (27%), Positives = 34/83 (40%)
Frame = +3
Query: 45 PVVSPPSPMIEVPLVDYPPSPTVEIPTVVSPPSPMVESSGEESSGEESSGEESSGEESSG 104
P +S PSP P PSP PT + PP+ S E + SS S+
Sbjct: 57 PTLSSPSPFAASP-----PSPRNPSPTTLPPPTTTNSPSKPEPPATSTPSATSS--TSAS 215
Query: 105 EESSGEASSGMGGSDEDSIPPPT 127
+ ++ S+ S + PPP+
Sbjct: 216 KTAASTPSTPSASSPPPTSPPPS 284
>AV777682
Length = 482
Score = 32.3 bits (72), Expect = 0.16
Identities = 24/76 (31%), Positives = 37/76 (48%), Gaps = 1/76 (1%)
Frame = +1
Query: 77 SPMVES-SGEESSGEESSGEESSGEESSGEESSGEASSGMGGSDEDSIPPPTVDDDVLPP 135
+PM ++ SG +S SS E +G+ S +E ++SGM D+D+ T D P
Sbjct: 94 NPMAQTMSGANASN--SSSEGPNGDSESNDEGQSLSNSGMNEEDDDA----TKDSQAADP 255
Query: 136 EQGEQGAQGGEEDLIQ 151
Q G G E+ +Q
Sbjct: 256 NQ--SGNHGQEQQGVQ 297
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.132 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,941,393
Number of Sequences: 28460
Number of extensions: 126026
Number of successful extensions: 1241
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 949
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1160
length of query: 448
length of database: 4,897,600
effective HSP length: 93
effective length of query: 355
effective length of database: 2,250,820
effective search space: 799041100
effective search space used: 799041100
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0318.7


