

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
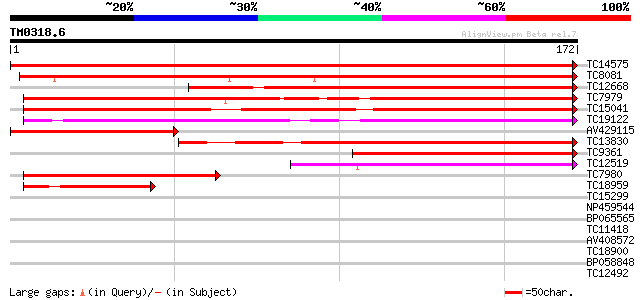
Query= TM0318.6
(172 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14575 similar to UP|AAQ84334 (AAQ84334) Zinc-finger protein, p... 356 1e-99
TC8081 similar to UP|AAQ84334 (AAQ84334) Zinc-finger protein, pa... 224 6e-60
TC12668 similar to UP|AAQ84334 (AAQ84334) Zinc-finger protein, p... 157 7e-40
TC7979 similar to UP|Q9SJM6 (Q9SJM6) At2g36320 protein (Zinc fin... 154 6e-39
TC15041 similar to UP|Q9SJM6 (Q9SJM6) At2g36320 protein (Zinc fi... 150 1e-37
TC19122 similar to UP|Q9LXI5 (Q9LXI5) Zinc finger-like protein, ... 143 2e-35
AV429115 108 5e-25
TC13830 similar to UP|Q94B40 (Q94B40) Zinc finger-like protein, ... 107 1e-24
TC9361 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (68%) 100 1e-22
TC12519 weakly similar to UP|BAC84565 (BAC84565) Zinc finger pro... 74 1e-14
TC7980 similar to UP|Q9SJM6 (Q9SJM6) At2g36320 protein (Zinc fin... 69 3e-13
TC18959 similar to UP|AAR07599 (AAR07599) Fiber protein Fb37, pa... 57 2e-09
TC15299 similar to UP|Q84MJ5 (Q84MJ5) Methylmalonate semialdehyd... 32 0.077
NP459544 ribosomal protein S18 [Lotus japonicus] 28 0.85
BP065565 27 1.4
TC11418 similar to GB|AAP12872.1|30017277|BT006223 At3g57480 {Ar... 26 3.2
AV408572 25 5.5
TC18900 similar to UP|BAC83916 (BAC83916) Methylmalonate semi-al... 25 5.5
BP058848 25 7.2
TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, parti... 25 9.4
>TC14575 similar to UP|AAQ84334 (AAQ84334) Zinc-finger protein, partial
(71%)
Length = 1080
Score = 356 bits (913), Expect = 1e-99
Identities = 172/172 (100%), Positives = 172/172 (100%)
Frame = +2
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN 60
MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN
Sbjct: 200 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVN 379
Query: 61 SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV 120
SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV
Sbjct: 380 SCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRV 559
Query: 121 GLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
GLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI
Sbjct: 560 GLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 715
>TC8081 similar to UP|AAQ84334 (AAQ84334) Zinc-finger protein, partial
(65%)
Length = 1222
Score = 224 bits (571), Expect = 6e-60
Identities = 108/175 (61%), Positives = 135/175 (76%), Gaps = 6/175 (3%)
Frame = +1
Query: 4 HDETGCQAP-ERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSC 62
H++T CQAP E P+LC+NNCGFFG AATMNMCSKC+KDM+LKQEQ KLAATS+ NI+N
Sbjct: 196 HEKTECQAPPESPMLCINNCGFFGSAATMNMCSKCHKDMVLKQEQAKLAATSIGNIMNGS 375
Query: 63 SNG---NGKQAITADAVNVRVEPVEVKAVTAQ--ISADSSSGESLEVKAKTGPSRCGTCR 117
S+ + + A +V++ V VE K +++Q S SGES E K K GP RC +C
Sbjct: 376 SSSIETEPTEPVVAASVDIPVISVEPKTISSQPVFGFGSGSGESGEEKPKDGPKRCSSCN 555
Query: 118 KRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
KRVGLTGF+C+CG++FCA+HRYSDKHDCPFDYR ++AIAKANPV+KA+KLDKI
Sbjct: 556 KRVGLTGFNCRCGHLFCAVHRYSDKHDCPFDYRTAARDAIAKANPVVKAEKLDKI 720
>TC12668 similar to UP|AAQ84334 (AAQ84334) Zinc-finger protein, partial
(37%)
Length = 513
Score = 157 bits (398), Expect = 7e-40
Identities = 78/118 (66%), Positives = 93/118 (78%)
Frame = +2
Query: 55 VENIVNSCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCG 114
VENIVN S +GKQA+T D V+V E K V +I DSSSGE+ EVKAKTGP+RC
Sbjct: 2 VENIVNGASGCSGKQALTVD---VQVGNAEFKTVCTEIREDSSSGENSEVKAKTGPNRCV 172
Query: 115 TCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
+CRKRVG TGFSCKCGN+FCA+ RY KHDCPFDY+ + +++IAKA+PVIKADKLDKI
Sbjct: 173 SCRKRVGFTGFSCKCGNLFCAIPRYFYKHDCPFDYQTLVRDSIAKAHPVIKADKLDKI 346
>TC7979 similar to UP|Q9SJM6 (Q9SJM6) At2g36320 protein (Zinc finger-like
protein), partial (73%)
Length = 1262
Score = 154 bits (390), Expect = 6e-39
Identities = 78/169 (46%), Positives = 112/169 (66%), Gaps = 1/169 (0%)
Frame = +2
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E C+APE LCVNNCGF G ATM++CSKCY+D+ LK++Q+ +++E ++S S+
Sbjct: 233 EEHRCEAPEGHRLCVNNCGFSGSPATMDLCSKCYRDIRLKEQQEASTKSTIETALSSSSS 412
Query: 65 -GNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLT 123
+ ++ + V + P KA TA IS ++G L+ P+RC TCRKRVGLT
Sbjct: 413 VAKPPSSTSSPSPAVDLAP-PAKAETASISL--TAGPILQAAQ---PNRCATCRKRVGLT 574
Query: 124 GFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
GF C+C FC HRY +KH C FD++ VG+EAIA+ NPV++A+KL +I
Sbjct: 575 GFKCRCEVTFCGAHRYPEKHACSFDFKTVGREAIARENPVVRAEKLRRI 721
>TC15041 similar to UP|Q9SJM6 (Q9SJM6) At2g36320 protein (Zinc finger-like
protein), partial (75%)
Length = 928
Score = 150 bits (378), Expect = 1e-37
Identities = 72/168 (42%), Positives = 103/168 (60%)
Frame = +1
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E GC+AP+ ILC NNCGFFG ATMN+CSKCY+D+ L + ++++ +++
Sbjct: 238 EEHGCKAPKGHILCANNCGFFGSPATMNLCSKCYRDVHLNDQDQAKTKSTIQTALST--- 408
Query: 65 GNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTG 124
+ A A VE +++ + + +V +RCGTCRKRVGLTG
Sbjct: 409 ------VAASAAASMPPAVESVPQPPALTSPEIASPAAQVT-----NRCGTCRKRVGLTG 555
Query: 125 FSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
F C+C FC HRY +KH C FD++AVG+E IA+ANPV++ADKL +I
Sbjct: 556 FRCRCEATFCGAHRYPEKHGCGFDFKAVGREEIARANPVVRADKLRRI 699
>TC19122 similar to UP|Q9LXI5 (Q9LXI5) Zinc finger-like protein, partial
(58%)
Length = 737
Score = 143 bits (360), Expect = 2e-35
Identities = 73/168 (43%), Positives = 99/168 (58%)
Frame = +3
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E CQAP LC NNCGFFG A ++CSKCY+D+ L ++Q A + + +
Sbjct: 207 EEHRCQAPR---LCANNCGFFGSPAMQDLCSKCYRDLQLNEQQSSSAKLVLNQTLAPPLS 377
Query: 65 GNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTG 124
+ + EPV V + A S+ E K GPSRC TCR+RVGLTG
Sbjct: 378 LSPAIPQPCSSSATTTEPVVV------VVAPSAPSE------KEGPSRCATCRRRVGLTG 521
Query: 125 FSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDKI 172
F C+CG + C HRY +KH+C FD++ +G+E IAKANPV+K +KL+KI
Sbjct: 522 FKCRCGLMLCGSHRYPEKHECGFDFKELGREQIAKANPVVKGEKLNKI 665
>AV429115
Length = 237
Score = 108 bits (270), Expect = 5e-25
Identities = 48/51 (94%), Positives = 50/51 (97%)
Frame = +1
Query: 1 MESHDETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLA 51
MESH+ETGCQAPER ILC+NNCGFFGRAATMNMCSKCYKDMLLKQEQDKLA
Sbjct: 85 MESHEETGCQAPERSILCINNCGFFGRAATMNMCSKCYKDMLLKQEQDKLA 237
>TC13830 similar to UP|Q94B40 (Q94B40) Zinc finger-like protein, partial
(37%)
Length = 648
Score = 107 bits (267), Expect = 1e-24
Identities = 54/121 (44%), Positives = 76/121 (62%)
Frame = +3
Query: 52 ATSVENIVNSCSNGNGKQAITADAVNVRVEPVEVKAVTAQISADSSSGESLEVKAKTGPS 111
A+S + ++N Q++ A V EP A S+ +S ++E GPS
Sbjct: 6 ASSAKQVLN--------QSLALPAPVVISEPA-----AASSSSVVASPRAVEEVKPNGPS 146
Query: 112 RCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVIKADKLDK 171
RC TCRKRVGLTGF C+CG C +HRY ++HDC FD++ +G++ IAKANPV+KA+KL+K
Sbjct: 147 RCVTCRKRVGLTGFKCRCGMTLCGVHRYPEQHDCEFDFKGLGRDQIAKANPVVKAEKLEK 326
Query: 172 I 172
I
Sbjct: 327 I 329
>TC9361 similar to UP|Q41123 (Q41123) PVPR3 protein, partial (68%)
Length = 781
Score = 100 bits (250), Expect = 1e-22
Identities = 41/68 (60%), Positives = 55/68 (80%)
Frame = +1
Query: 105 KAKTGPSRCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHDCPFDYRAVGQEAIAKANPVI 164
+AK +RC CR+RVGLTGF C+CG +FCA HRYSD+HDC +DY+A G+EAIA+ NPV+
Sbjct: 337 EAKRAVNRCSGCRRRVGLTGFRCRCGELFCADHRYSDRHDCSYDYKAAGREAIARENPVV 516
Query: 165 KADKLDKI 172
+A K+ K+
Sbjct: 517 RAAKIIKL 540
>TC12519 weakly similar to UP|BAC84565 (BAC84565) Zinc finger protein-like,
partial (42%)
Length = 686
Score = 74.3 bits (181), Expect = 1e-14
Identities = 35/88 (39%), Positives = 52/88 (58%), Gaps = 1/88 (1%)
Frame = +2
Query: 86 KAVTAQISADSSSGESLEV-KAKTGPSRCGTCRKRVGLTGFSCKCGNVFCAMHRYSDKHD 144
+A TA +A + L K + RC +CRK+VGLT C+CG V+C HR + H+
Sbjct: 23 EAGTASTAAPADEVPDLPARKVQKDKRRCFSCRKKVGLTALPCRCGYVYCGAHRMATDHN 202
Query: 145 CPFDYRAVGQEAIAKANPVIKADKLDKI 172
C FDY+ + +A ANP++ A K++KI
Sbjct: 203 CDFDYKTHARTVLAGANPLVAAVKVNKI 286
>TC7980 similar to UP|Q9SJM6 (Q9SJM6) At2g36320 protein (Zinc finger-like
protein), partial (36%)
Length = 670
Score = 69.3 bits (168), Expect = 3e-13
Identities = 29/60 (48%), Positives = 44/60 (73%)
Frame = +2
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSCSN 64
+E C+APE LCVNNCGF G ATM++CSKCY+D+ LK++Q+ +++E ++S S+
Sbjct: 419 EEHRCEAPEGHRLCVNNCGFSGSPATMDLCSKCYRDIRLKEQQEASTKSTIETALSSSSS 598
>TC18959 similar to UP|AAR07599 (AAR07599) Fiber protein Fb37, partial (28%)
Length = 364
Score = 57.0 bits (136), Expect = 2e-09
Identities = 24/40 (60%), Positives = 30/40 (75%)
Frame = +3
Query: 5 DETGCQAPERPILCVNNCGFFGRAATMNMCSKCYKDMLLK 44
+E CQAP + C NNCGFFG AAT N+CS+CY+D+ LK
Sbjct: 252 EEHRCQAP---LFCANNCGFFGSAATENLCSRCYRDLQLK 362
>TC15299 similar to UP|Q84MJ5 (Q84MJ5) Methylmalonate semialdehyde
dehydrogenase (Fragment), partial (31%)
Length = 570
Score = 31.6 bits (70), Expect = 0.077
Identities = 21/57 (36%), Positives = 27/57 (46%), Gaps = 5/57 (8%)
Frame = +3
Query: 36 KCYKD-----MLLKQEQDKLAATSVENIVNSCSNGNGKQAITADAVNVRVEPVEVKA 87
+CYK+ +LL E D L NIVN GNG TA V+ R E++A
Sbjct: 33 ECYKEEIFGPVLLLMESDSLEKAI--NIVNENKYGNGASIFTASGVSARKFQTEIEA 197
>NP459544 ribosomal protein S18 [Lotus japonicus]
Length = 315
Score = 28.1 bits (61), Expect = 0.85
Identities = 10/34 (29%), Positives = 20/34 (58%)
Frame = -2
Query: 29 ATMNMCSKCYKDMLLKQEQDKLAATSVENIVNSC 62
A ++ S C+ +L+K +DK+ A + ++N C
Sbjct: 275 AVNSLFSNCFFSLLIKGNEDKIRACFIAIVINRC 174
>BP065565
Length = 536
Score = 27.3 bits (59), Expect = 1.4
Identities = 13/29 (44%), Positives = 13/29 (44%), Gaps = 2/29 (6%)
Frame = -2
Query: 111 SRCGTCRKRVGLTGFSCK--CGNVFCAMH 137
S CG C G G CK C N CA H
Sbjct: 217 SLCGLCNNYFGTIG*RCKNICTNKMCAKH 131
>TC11418 similar to GB|AAP12872.1|30017277|BT006223 At3g57480 {Arabidopsis
thaliana;}, partial (81%)
Length = 994
Score = 26.2 bits (56), Expect = 3.2
Identities = 11/29 (37%), Positives = 15/29 (50%), Gaps = 1/29 (3%)
Frame = +1
Query: 118 KRVGLTGFSC-KCGNVFCAMHRYSDKHDC 145
K V F+C +C V+C HR +H C
Sbjct: 136 KLVDFLPFTCDRCNQVYCLEHRSYIRHQC 222
Score = 24.6 bits (52), Expect = 9.4
Identities = 20/79 (25%), Positives = 31/79 (38%), Gaps = 7/79 (8%)
Frame = +1
Query: 75 AVNVRVEPVEVKAVT----AQISADSSSGESLEVKAKTGPSRCGTCRKRVGLTGFSCKCG 130
A VR+ P + +T D S+ E + K K + C K + + + KC
Sbjct: 271 AKGVRLVPDQDPNITWENHVNTDCDPSNYEKVTKKKKCPAAGC----KEILVFSNTIKCR 438
Query: 131 NVF---CAMHRYSDKHDCP 146
+ C HR+ H CP
Sbjct: 439 DCMLDHCLKHRFGPDHKCP 495
>AV408572
Length = 359
Score = 25.4 bits (54), Expect = 5.5
Identities = 16/57 (28%), Positives = 21/57 (36%), Gaps = 15/57 (26%)
Frame = +1
Query: 113 CGTCRKRVGLTGFSCKCGNVF---------------CAMHRYSDKHDCPFDYRAVGQ 154
C T + G +GF C+C + F + YS C FDY A Q
Sbjct: 64 CTTVNLQNGSSGFRCRCNDGFQGDGFANGAGCWKG*LRLKIYSS*LKCMFDYTATSQ 234
>TC18900 similar to UP|BAC83916 (BAC83916) Methylmalonate semi-aldehyde
dehydrogenase, partial (42%)
Length = 809
Score = 25.4 bits (54), Expect = 5.5
Identities = 20/63 (31%), Positives = 28/63 (43%), Gaps = 5/63 (7%)
Frame = +1
Query: 30 TMNMCSKCYKD-----MLLKQEQDKLAATSVENIVNSCSNGNGKQAITADAVNVRVEPVE 84
T NM +CYK+ +LL E D L +I+N GNG T V R +
Sbjct: 346 TANM--ECYKEEIFGPVLLFMEADSLEEAI--DIINKNQYGNGASIFTTSGVAARKFQTD 513
Query: 85 VKA 87
++A
Sbjct: 514 IEA 522
>BP058848
Length = 485
Score = 25.0 bits (53), Expect = 7.2
Identities = 10/19 (52%), Positives = 11/19 (57%)
Frame = -3
Query: 17 LCVNNCGFFGRAATMNMCS 35
LC NC FF MN+CS
Sbjct: 102 LCTVNCHFFPIFTLMNLCS 46
>TC12492 weakly similar to UP|AAS53933 (AAS53933) AFR562Cp, partial (7%)
Length = 922
Score = 24.6 bits (52), Expect = 9.4
Identities = 10/23 (43%), Positives = 12/23 (51%)
Frame = -3
Query: 20 NNCGFFGRAATMNMCSKCYKDML 42
N FFG T+N CS YK +
Sbjct: 446 NTIRFFGVPLTLNSCSLIYKQFI 378
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.131 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,225,306
Number of Sequences: 28460
Number of extensions: 43765
Number of successful extensions: 235
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 227
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 230
length of query: 172
length of database: 4,897,600
effective HSP length: 84
effective length of query: 88
effective length of database: 2,506,960
effective search space: 220612480
effective search space used: 220612480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0318.6


