

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0318.13
(643 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
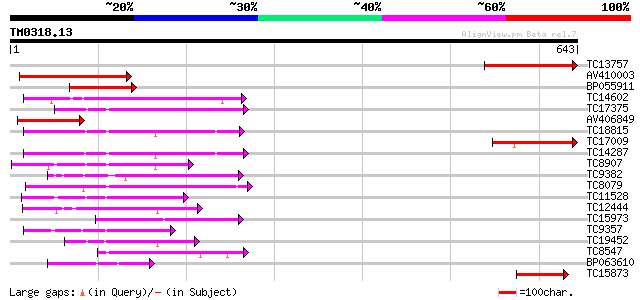
Score E
Sequences producing significant alignments: (bits) Value
TC13757 similar to UP|Q9LT35 (Q9LT35) Kinase-like protein, parti... 210 6e-55
AV410003 192 2e-49
BP055911 122 1e-28
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 118 3e-27
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 116 1e-26
AV406849 114 6e-26
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 110 5e-25
TC17009 similar to UP|Q9LT35 (Q9LT35) Kinase-like protein, parti... 109 1e-24
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 108 3e-24
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 95 4e-20
TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein ... 93 1e-19
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 90 1e-18
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 89 2e-18
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 87 6e-18
TC15973 homologue to GB|AAL15278.1|16323087|AY057647 AT3g01490/F... 86 1e-17
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 82 3e-16
TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, parti... 82 3e-16
TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated pro... 79 2e-15
BP063610 79 3e-15
TC15873 similar to UP|Q9LXP3 (Q9LXP3) Protein kinase-like protei... 74 6e-14
>TC13757 similar to UP|Q9LT35 (Q9LT35) Kinase-like protein, partial (12%)
Length = 500
Score = 210 bits (534), Expect = 6e-55
Identities = 105/105 (100%), Positives = 105/105 (100%)
Frame = +3
Query: 539 KDNETAKENIICTTQKDENLVGVDQTPSDTTMDDGDETMRDSPCQQRADALESLLELCAQ 598
KDNETAKENIICTTQKDENLVGVDQTPSDTTMDDGDETMRDSPCQQRADALESLLELCAQ
Sbjct: 3 KDNETAKENIICTTQKDENLVGVDQTPSDTTMDDGDETMRDSPCQQRADALESLLELCAQ 182
Query: 599 LLKQDKLEELAGVLKPFGKEAVSSRETAIWLTKSLMSAQKFNSET 643
LLKQDKLEELAGVLKPFGKEAVSSRETAIWLTKSLMSAQKFNSET
Sbjct: 183 LLKQDKLEELAGVLKPFGKEAVSSRETAIWLTKSLMSAQKFNSET 317
>AV410003
Length = 428
Score = 192 bits (487), Expect = 2e-49
Identities = 86/127 (67%), Positives = 112/127 (87%)
Frame = +3
Query: 12 KMEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLN 71
+M+ YE++EQIGRGAFG+A LV HK+E+K+YVLKKIRLA+QTE+ +R+AHQEM LIA++
Sbjct: 48 RMDQYEIMEQIGRGAFGAAILVNHKAERKKYVLKKIRLARQTERCRRSAHQEMALIARIQ 227
Query: 72 NPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDY 131
+PYIV++K+AWVEK +CIVTGYCEGGDMAE +KK G++ PEEK+CKW TQLL+AV+Y
Sbjct: 228 HPYIVEFKEAWVEKGCYVCIVTGYCEGGDMAELMKKQNGAYIPEEKLCKWFTQLLLAVEY 407
Query: 132 LHSNRVI 138
LHSN V+
Sbjct: 408 LHSNYVL 428
>BP055911
Length = 373
Score = 122 bits (307), Expect = 1e-28
Identities = 52/76 (68%), Positives = 68/76 (89%)
Frame = +2
Query: 68 AKLNNPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLI 127
A++ +PYIV++K+AWVEK +CIVTGYCEGGDMA +KK+ G++FPEEK+CKW+TQLL+
Sbjct: 68 ARIQHPYIVEFKEAWVEKGCFVCIVTGYCEGGDMAXLMKKSNGAYFPEEKLCKWITQLLL 247
Query: 128 AVDYLHSNRVIHRDLK 143
AV+YLHSN V+HRDLK
Sbjct: 248 AVEYLHSNFVLHRDLK 295
Score = 37.0 bits (84), Expect = 0.010
Identities = 16/41 (39%), Positives = 30/41 (73%)
Frame = +3
Query: 46 KIRLAKQTEKFKRTAHQEMNLIAKLNNPYIVDYKDAWVEKE 86
KIRLA+QTE+ +R+A+QEM L+ + + P +++ + + K+
Sbjct: 3 KIRLARQTERCRRSAYQEMALMLEFSTPTLLNLRRHGLRKD 125
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 118 bits (295), Expect = 3e-27
Identities = 77/260 (29%), Positives = 130/260 (49%), Gaps = 7/260 (2%)
Frame = +2
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLK---KIRLAKQTEKF-KRTAHQEMNLIAKLN 71
Y++ E+IGRG FG+ F H + +K K LA T++ + M+L++
Sbjct: 68 YQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLLSP-- 241
Query: 72 NPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDY 131
+P I+ D + E +D + +V C+ + + I A G+ PE + + QLL AV +
Sbjct: 242 HPNILQIFDVF-EDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAH 418
Query: 132 LHSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAEDLTSSVVGTPNYMCPEILADIP 191
H V HRD+K N+ +++L DFG A+ S VVGTP Y+ PE+L
Sbjct: 419 CHRLGVAHRDVKPDNVLFGGGGDLKLADFGSAEWFGDGRRMSGVVGTPYYVAPEVLMGRE 598
Query: 192 YGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVY---SSTLKQIIK 248
YG K D+WS G ++ + + P F A + + R ++ ++ S K +++
Sbjct: 599 YGEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAVIRGNLRFPSRIFRNVSPAAKDLLR 778
Query: 249 SMLRKNPEHRPTAAELLRHP 268
M+ ++P +R +A + LRHP
Sbjct: 779 KMICRDPSNRISAEQALRHP 838
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 116 bits (290), Expect = 1e-26
Identities = 69/223 (30%), Positives = 121/223 (53%), Gaps = 2/223 (0%)
Frame = +3
Query: 51 KQTEKFKRTAHQEMNLIAKLNNPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARG 110
K +++ + +QE+NL+ + ++P IV Y + + +E + + Y GG + + +++
Sbjct: 45 KTSKECLKQLNQEINLLNQFSHPNIVQYYGSELGEES-LSVYLEYVSGGSIHKLLQEYGA 221
Query: 111 SFFPEEKVCKWLTQLLIAVDYLHSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAED 170
F E + + Q++ + YLHS +HRD+K +NI + + I+L DFG++K +N+
Sbjct: 222 --FKEPVIQNYTRQIVSGLAYLHSRNTVHRDIKGANILVDPNGEIKLADFGMSKHINSAA 395
Query: 171 LTSSVVGTPNYMCPEILADI-PYGYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINR 229
S G+P +M PE++ + YG D+ SLGC + E+A +P + + I KI
Sbjct: 396 SMLSFKGSPYWMAPEVVMNTNGYGLPVDISSLGCTILEMATSKPPWSQFEGVAAIFKIGN 575
Query: 230 SSISP-LPIVYSSTLKQIIKSMLRKNPEHRPTAAELLRHPHLQ 271
S P +P S K IK L+++P RPTA LL HP ++
Sbjct: 576 SKDMPEIPEHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFIR 704
>AV406849
Length = 259
Score = 114 bits (284), Expect = 6e-26
Identities = 51/76 (67%), Positives = 67/76 (88%)
Frame = +1
Query: 10 SKKMEDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAK 69
++KME YEV+EQIG+G+F SA LV HK E K+YVLKKIRLA+QT++ +R+AHQEM LI+K
Sbjct: 22 AEKMEQYEVLEQIGKGSFASALLVRHKHENKKYVLKKIRLARQTDRTRRSAHQEMELISK 201
Query: 70 LNNPYIVDYKDAWVEK 85
+ NP+IV+YKD+WVEK
Sbjct: 202VRNPFIVEYKDSWVEK 249
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 110 bits (276), Expect = 5e-25
Identities = 69/256 (26%), Positives = 131/256 (50%), Gaps = 5/256 (1%)
Frame = +2
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAK-QTEKFKRTAHQEMNLIAKLNNPY 74
YE+ +G+G + + + +K + A+ + E +E++++ + +P
Sbjct: 38 YEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMRLVRHPN 217
Query: 75 IVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHS 134
IV+ K+ K +I + Y GG++ + K + ++ ++ QL+ AVDY HS
Sbjct: 218 IVNLKEVMATKT-KIFFIMEYIRGGELFAKVAKGK---LKDDLARRYFQQLISAVDYCHS 385
Query: 135 NRVIHRDLKCSNIFLTKDNNIRLGDFGLA---KRLNAEDLTSSVVGTPNYMCPEILADIP 191
V HRDLK N+ L ++ N+++ DFGL+ ++L + L + GTP Y+ PE+L
Sbjct: 386 RGVSHRDLKPENLLLDENENLKVSDFGLSGLPEQLRQDGLLHTQCGTPAYVAPEVLRKKG 565
Query: 192 Y-GYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSM 250
Y G+K+D WS G ++ + A F+ ++ + NK+ R+ P +S K++I +
Sbjct: 566 YDGFKTDTWSCGVILYALLAGCLPFQHENLMTMYNKVLRAEFQ-FPPWFSPESKKLISKI 742
Query: 251 LRKNPEHRPTAAELLR 266
L +P R T + ++R
Sbjct: 743 LVADPNRRITISSIMR 790
>TC17009 similar to UP|Q9LT35 (Q9LT35) Kinase-like protein, partial (12%)
Length = 542
Score = 109 bits (273), Expect = 1e-24
Identities = 62/105 (59%), Positives = 75/105 (71%), Gaps = 9/105 (8%)
Frame = +2
Query: 548 IICTTQKDENLVGV-DQTPSDTTMD------DGDETMR--DSPCQQRADALESLLELCAQ 598
I C+ QK+ + V D+TP+D ++ GD+T R D C RADALESLLELCAQ
Sbjct: 8 ISCSMQKENDKANVIDKTPNDISLSRHVLVGGGDKTKRRVDISCPPRADALESLLELCAQ 187
Query: 599 LLKQDKLEELAGVLKPFGKEAVSSRETAIWLTKSLMSAQKFNSET 643
LLKQ KLEELA VL+PFG++ VSSRETAIWLTKSL+S+ N ET
Sbjct: 188 LLKQGKLEELAAVLRPFGEDVVSSRETAIWLTKSLISSPTINPET 322
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 108 bits (270), Expect = 3e-24
Identities = 68/261 (26%), Positives = 136/261 (52%), Gaps = 5/261 (1%)
Frame = +3
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAK-QTEKFKRTAHQEMNLIAKLNNPY 74
YE+ +G+G F + + + + +K I+ K + ++ + +E++++ + +P+
Sbjct: 432 YEMGRVLGQGNFAKVYHGRNLATNENVAIKVIKKEKLKKDRLVKQIKREVSVMRLVRHPH 611
Query: 75 IVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHS 134
IV+ K+ K +I +V Y +GG++ + K + E+ K+ QL+ AVD+ HS
Sbjct: 612 IVELKEVMATK-GKIFLVMEYVKGGELFTKVNKGK---LNEDDARKYFQQLISAVDFCHS 779
Query: 135 NRVIHRDLKCSNIFLTKDNNIRLGDFGLA---KRLNAEDLTSSVVGTPNYMCPEILADIP 191
V HRDLK N+ L ++ ++++ DFGL+ ++ + + + GTP Y+ PE+L
Sbjct: 780 RGVTHRDLKPENLLLDENEDLKVSDFGLSALPEQRRDDGMLVTPCGTPAYVAPEVLKKKG 959
Query: 192 Y-GYKSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSM 250
Y G K+D+WS G ++ + + F+ ++ + K ++ P S K +I ++
Sbjct: 960 YDGSKADIWSCGVILYALLSGYLPFQGENVMRIYRKAFKAEYE-FPEWISPQAKNLISNL 1136
Query: 251 LRKNPEHRPTAAELLRHPHLQ 271
L +PE R + E++ P Q
Sbjct: 1137LVADPEKRYSIPEIISDPWFQ 1199
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 94.7 bits (234), Expect = 4e-20
Identities = 61/213 (28%), Positives = 113/213 (52%), Gaps = 7/213 (3%)
Frame = +3
Query: 3 TENSDMRSKKMEDYEVIEQIGRGAFGSAFLVLHKSEKKRY---VLKKIRLAKQTEKFKRT 59
T + R+ YE+ + +G+G F + + + V+KK RL K E+ +
Sbjct: 303 TMRAAKRNILFNKYEIGKILGQGNFAKVYHGRNMETNESVAIKVIKKERLKK--ERLVKQ 476
Query: 60 AHQEMNLIAKLNNPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVC 119
+E++++ + +P+IV+ K+ K +I +V Y +GG++ + K + E
Sbjct: 477 IKREVSVMRLVRHPHIVELKEVMATKT-KIFMVVEYVKGGELFAKLTKGK---MTEVAAR 644
Query: 120 KWLTQLLIAVDYLHSNRVIHRDLKCSNIFLTKDNNIRLGDFGLA---KRLNAEDLTSSVV 176
K+ QL+ AVD+ HS V HRDLK N+ L + ++++ DFGL+ ++ ++ + +
Sbjct: 645 KYFQQLISAVDFCHSRGVTHRDLKPENLLLDDNEDLKVSDFGLSSLPEQRRSDGMLLTPC 824
Query: 177 GTPNYMCPEILADIPY-GYKSDMWSLGCCMFEI 208
GTP Y+ PE+L Y G K+D+WS G ++ +
Sbjct: 825 GTPAYVAPEVLKKKGYDGSKADIWSCGVILYAL 923
>TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein kinase Abl
(D-ash) , partial (5%)
Length = 1197
Score = 93.2 bits (230), Expect = 1e-19
Identities = 59/230 (25%), Positives = 117/230 (50%), Gaps = 7/230 (3%)
Frame = +2
Query: 43 VLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYIVDYKDAWVEKEDRICIVTGYCEGGDMA 102
VLK R++ T+ K A QE+ ++ K+ + +V + A + +CIVT + G +
Sbjct: 29 VLKPERIS--TDMLKEFA-QEVYIMRKIRHKNVVQFIGACT-RTPNLCIVTEFMSRGSLY 196
Query: 103 ENIKKARGSFFPEEKVCKWLTQLLIAVD------YLHSNRVIHRDLKCSNIFLTKDNNIR 156
+ + K RG F K + L +A+D YLH N +IHRDLK N+ + ++ ++
Sbjct: 197 DFLHKQRGVF-------KLPSLLKVAIDVSKGMNYLHQNNIIHRDLKTGNLLMDENELVK 355
Query: 157 LGDFGLAKRLNAEDLTSSVVGTPNYMCPEILADIPYGYKSDMWSLGCCMFEIAAHQ-PAF 215
+ DFG+A+ + + ++ GT +M PE++ PY K+D++S G ++E+ + P
Sbjct: 356 VADFGVARVITQSGVMTAETGTYRWMAPEVIEHKPYDQKADVFSFGIALWELLTGELPYS 535
Query: 216 RAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSMLRKNPEHRPTAAELL 265
+ + + + +P L ++++ +++P RP +E++
Sbjct: 536 YLTPLQAAVGVVQKGLRPSIPKNTHPRLSELLQRCWKQDPIERPAFSEII 685
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 90.1 bits (222), Expect = 1e-18
Identities = 67/261 (25%), Positives = 118/261 (44%), Gaps = 4/261 (1%)
Frame = +3
Query: 19 IEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYIVDY 78
I IGRGA+G +L+ + +KKI A + +E+ L+ L++ ++
Sbjct: 300 IMPIGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLREIKLLRHLDHENVIAL 479
Query: 79 KDAW---VEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSN 135
+D + +E +T D+ + I+ +G EE +L Q+L + Y+HS
Sbjct: 480 RDVIPPPLRREFTDVYITTELMDTDLHQIIRSNQG--LSEEHCQYFLYQVLRGLKYIHSA 653
Query: 136 RVIHRDLKCSNIFLTKDNNIRLGDFGLAKRLNAEDLTSSVVGTPNYMCPEILAD-IPYGY 194
+IHRDLK SN+ L + ++++ DFGLA+ D + V T Y PE+L + Y
Sbjct: 654 NIIHRDLKPSNLLLNSNCDLKIIDFGLARPTVENDFMTEYVVTRWYRAPELLLNSSDYTS 833
Query: 195 KSDMWSLGCCMFEIAAHQPAFRAPDMAGLINKINRSSISPLPIVYSSTLKQIIKSMLRKN 254
D+WS+GC E+ +P F D + + +P ++ +R+
Sbjct: 834 AIDVWSVGCIFMELMNKKPLFPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVRRYIRQL 1013
Query: 255 PEHRPTAAELLRHPHLQPYVL 275
P++ P PH+ P +
Sbjct: 1014PQY-PRQPLTKVFPHVHPMAM 1073
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 89.0 bits (219), Expect = 2e-18
Identities = 63/192 (32%), Positives = 93/192 (47%), Gaps = 3/192 (1%)
Frame = +3
Query: 14 EDYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNP 73
E YE ++++G G FG A L K+ + +K I K K +E+ L +P
Sbjct: 72 ERYEPLKELGSGNFGVARLARDKNTGELVAVKYIERGK---KIDENVQREIINHRSLRHP 242
Query: 74 YIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLH 133
I+ +K+ + + IV Y GG++ E I A F E++ + QL+ V Y H
Sbjct: 243 NIIRFKEVLLTPT-HLAIVLEYASGGELFERICSA--GRFSEDEARYFFQQLISGVSYCH 413
Query: 134 SNRVIHRDLKCSNIFLTKDNNIRLG--DFGLAKRLNAEDLTSSVVGTPNYMCPEILADIP 191
S + HRDLK N L + + RL DFG +K S VGTP Y+ PE+L+
Sbjct: 414 SMEICHRDLKLENTLLDGNPSPRLKICDFGYSKSAILHSQPKSTVGTPAYIAPEVLSRKE 593
Query: 192 Y-GYKSDMWSLG 202
Y G +D+WS G
Sbjct: 594 YDGKVADVWSCG 629
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 87.4 bits (215), Expect = 6e-18
Identities = 64/213 (30%), Positives = 106/213 (49%), Gaps = 9/213 (4%)
Frame = +2
Query: 15 DYEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAK---QTEKFKRTAHQEMNLIAKLN 71
DY ++E++G GA + + + + +K + L + E +R A Q M+LI N
Sbjct: 86 DYNLLEEVGYGASATVYRAVFLPRDEEVAVKCVDLDRCNANLEDIRREA-QTMSLIDHRN 262
Query: 72 NPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDY 131
+V ++V + R+ +V + G +K A F EE + L + L A++Y
Sbjct: 263 ---VVRAHWSFVV-DRRLWVVMPFMAQGSCLHLMKVAYPDGFEEEAIGSILKETLKALEY 430
Query: 132 LHSNRVIHRDLKCSNIFLTKDNNIRLGDFGLAKRL----NAEDLTSSVVGTPNYMCPEIL 187
LH + IHRD+K NI L D ++L DFG++ + + ++ VGTP +M PE+L
Sbjct: 431 LHRHGHIHRDVKAGNILLGSDGQVKLADFGVSASMFDAGERQRNRNTFVGTPCWMAPEVL 610
Query: 188 -ADIPYGYKSDMWSLGCCMFEIA-AHQPAFRAP 218
Y +K+D+WS G E+A H P + P
Sbjct: 611 QPGTGYNFKADVWSFGITALELAHGHAPFSKYP 709
>TC15973 homologue to GB|AAL15278.1|16323087|AY057647 AT3g01490/F4P13_4
{Arabidopsis thaliana;}, partial (49%)
Length = 944
Score = 86.3 bits (212), Expect = 1e-17
Identities = 50/171 (29%), Positives = 85/171 (49%), Gaps = 3/171 (1%)
Frame = +3
Query: 98 GGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSNRVIHRDLKCSNIFLTKDNNIRL 157
GG + + K R + V + L + YLHS +++HRD+K N+ L K +++
Sbjct: 3 GGTLKSYLIKNRRRKLAFKVVIQLALDLARGLSYLHSQKIVHRDVKTENMLLDKTRTVKI 182
Query: 158 GDFGLAK--RLNAEDLTSSVVGTPNYMCPEILADIPYGYKSDMWSLGCCMFEIAAHQPAF 215
DFG+A+ N D+T GT YM PE+L PY K D++S G C++EI +
Sbjct: 183 ADFGVARVEASNPNDMTGE-TGTLGYMAPEVLNGNPYNRKCDVYSFGICLWEIYCCDMPY 359
Query: 216 RAPDMAGLINKINRSSISP-LPIVYSSTLKQIIKSMLRKNPEHRPTAAELL 265
+ + + + R ++ P +P S+L ++K P+ RP E++
Sbjct: 360 PDLSFSEITSAVVRQNLRPEIPRCCPSSLANVMKKCWDATPDKRPEMDEVV 512
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 81.6 bits (200), Expect = 3e-16
Identities = 55/175 (31%), Positives = 87/175 (49%), Gaps = 2/175 (1%)
Frame = +1
Query: 16 YEVIEQIGRGAFGSAFLVLHKSEKKRYVLKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYI 75
Y+++ IG G FG A L+ K K+ +K I ++ +K +E+ L +P I
Sbjct: 361 YDLVRDIGSGNFGVARLMQDKQTKELVAVKYI---ERGDKIDENVKREIINHRSLRHPNI 531
Query: 76 VDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSN 135
V +K+ + + IV Y GG++ E I A F E++ + QL+ V Y H+
Sbjct: 532 VRFKEV-ILTPTHLAIVMEYASGGELFERICNA--GRFTEDEARFFFQQLISGVSYCHAM 702
Query: 136 RVIHRDLKCSNIFL--TKDNNIRLGDFGLAKRLNAEDLTSSVVGTPNYMCPEILA 188
+V HRDLK N L + +++ DFG +K S VGTP Y+ PE+L+
Sbjct: 703 QVCHRDLKLENTLLDGSPTPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLS 867
>TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, partial (41%)
Length = 562
Score = 81.6 bits (200), Expect = 3e-16
Identities = 51/162 (31%), Positives = 91/162 (55%), Gaps = 9/162 (5%)
Frame = +3
Query: 63 EMNLIAKLNNPYIVDYKDAWVEKEDRICIVTGYCEGGDMAENIKKARGSFFPEEKVCKWL 122
E+NL+ KL +P IV + A E++ + ++T Y GGD+ + +K+ +GS P + +
Sbjct: 48 EVNLLVKLRHPNIVQFLGAVTERKP-LMLITEYLRGGDLHQYLKE-KGSLSPSTAI-NFS 218
Query: 123 TQLLIAVDYLHS--NRVIHRDLKCSNIFLTKD--NNIRLGDFGLAKRL---NAEDL--TS 173
++ + YLH+ N +IHRDLK N+ L +++++GDFGL+K + N+ D+ +
Sbjct: 219 MDIVRGMAYLHNEPNVIIHRDLKPRNVLLVNSSADHLKVGDFGLSKLITVQNSHDVYKMT 398
Query: 174 SVVGTPNYMCPEILADIPYGYKSDMWSLGCCMFEIAAHQPAF 215
G+ YM PE+ Y K D++S ++E+ +P F
Sbjct: 399 GETGSYRYMAPEVFKHRKYDKKVDVYSFAMILYEMLEGEPPF 524
>TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated protein
kinase homolog MMK1 (MAP kinase MSK7) (MAP kinase ERK1)
, partial (71%)
Length = 1098
Score = 79.3 bits (194), Expect = 2e-15
Identities = 57/201 (28%), Positives = 93/201 (45%), Gaps = 30/201 (14%)
Frame = +1
Query: 100 DMAENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSNRVIHRDLKCSNIFLTKDNNIRLGD 159
D+ + I+ +G EE +L Q+L + Y+HS V+HRDLK SN+ L + ++++ D
Sbjct: 94 DLHQIIRSNQG--LSEEHCQYFLYQILRGLKYIHSANVLHRDLKPSNLLLNANCDLKICD 267
Query: 160 FGLAKRLNAEDLTSSVVGTPNYMCPEILAD-IPYGYKSDMWSLGCCMFEIAAHQPAF--- 215
FGLA+ + D + V T Y PE+L + Y D+WS+GC E+ +P F
Sbjct: 268 FGLARVTSETDFMTEYVVTRWYRAPELLLNSSDYTAAIDVWSVGCIFMELMDRKPLFPGR 447
Query: 216 ---------------RAPDMAGLINKINRSSISPLPIVYSSTLKQ-----------IIKS 249
+ D G +N+ + I LP + ++ +++
Sbjct: 448 DHVHQLRLLMELIGTPSEDDLGFLNENAKRYIRQLPPYRRQSFQEKFPQVHPEAIDLVEK 627
Query: 250 MLRKNPEHRPTAAELLRHPHL 270
ML +P R T E L HP+L
Sbjct: 628 MLTFDPRKRITVEEALAHPYL 690
>BP063610
Length = 461
Score = 78.6 bits (192), Expect = 3e-15
Identities = 47/122 (38%), Positives = 70/122 (56%), Gaps = 1/122 (0%)
Frame = -3
Query: 44 LKKIRLAKQTEKFKRTAHQEMNLIAKLNNPYIVDYKDAWVEKE-DRICIVTGYCEGGDMA 102
LKK+ + K+ E F T+ +E+N++ ++PYIVD K+ V D I +V Y E D+
Sbjct: 441 LKKVXMEKEKEGFPLTSLREINILLSFHHPYIVDVKEVVVGSSLDSIFMVMEYMEH-DLK 265
Query: 103 ENIKKARGSFFPEEKVCKWLTQLLIAVDYLHSNRVIHRDLKCSNIFLTKDNNIRLGDFGL 162
++ + F E C + QLL V YLH N V+HRDLK SN+ L +++ DFGL
Sbjct: 264 GLMESMKQPFSQSEVKCLMI-QLLEGVKYLHDNWVLHRDLKTSNLLLNNRGELKICDFGL 88
Query: 163 AK 164
A+
Sbjct: 87 AR 82
>TC15873 similar to UP|Q9LXP3 (Q9LXP3) Protein kinase-like protein, partial
(7%)
Length = 522
Score = 74.3 bits (181), Expect = 6e-14
Identities = 39/59 (66%), Positives = 46/59 (77%)
Frame = +1
Query: 575 ETMRDSPCQQRADALESLLELCAQLLKQDKLEELAGVLKPFGKEAVSSRETAIWLTKSL 633
ET+ QRA+ALE LLEL A LL+Q++LEEL+ VLKPFGK+ VS RETAIWL KSL
Sbjct: 58 ETLDVKSSTQRAEALEGLLELSADLLQQNRLEELSIVLKPFGKDKVSPRETAIWLAKSL 234
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.129 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,390,072
Number of Sequences: 28460
Number of extensions: 142522
Number of successful extensions: 795
Number of sequences better than 10.0: 188
Number of HSP's better than 10.0 without gapping: 748
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 755
length of query: 643
length of database: 4,897,600
effective HSP length: 96
effective length of query: 547
effective length of database: 2,165,440
effective search space: 1184495680
effective search space used: 1184495680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0318.13


