

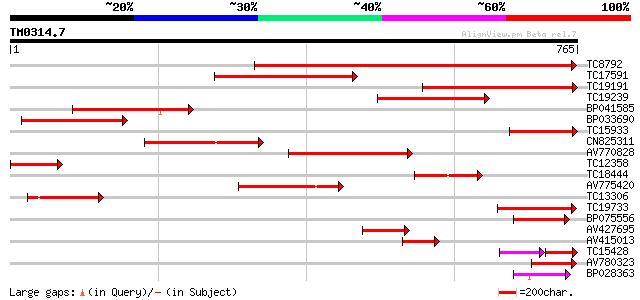
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0314.7
(765 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8792 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosid... 453 e-128
TC17591 similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (... 388 e-108
TC19191 similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragme... 352 1e-97
TC19239 homologue to UP|Q9ZU04 (Q9ZU04) Beta-glucosidase (Fragm... 307 4e-84
BP041585 268 2e-72
BP033690 226 8e-60
TC15933 weakly similar to UP|BAC98299 (BAC98299) LEXYL2 protein ... 180 9e-46
CN825311 177 6e-45
AV770828 174 6e-44
TC12358 weakly similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, pa... 149 2e-36
TC18444 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial... 131 5e-31
AV775420 130 6e-31
TC13306 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (12%) 119 2e-27
TC19733 weakly similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11,... 97 7e-21
BP075556 75 5e-14
AV427695 74 7e-14
AV415013 69 3e-12
TC15428 44 6e-11
AV780323 61 6e-10
BP028363 59 2e-09
>TC8792 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosidase
{Prunus persica;} , partial (90%)
Length = 1636
Score = 453 bits (1166), Expect = e-128
Identities = 230/437 (52%), Positives = 297/437 (67%), Gaps = 3/437 (0%)
Frame = +3
Query: 331 LGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQE 390
L +T+ A+K GL+ E +N A++N MRLG FDG PS PYG LGP+DVCT + E
Sbjct: 3 LALHTDAAIKNGLITENDLNLALANLITVQMRLGMFDGEPSAHPYGNLGPRDVCTPAHNE 182
Query: 391 LAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQG 450
LA EAARQGIVLL+N +LPLS +++ VIGPN++ T MIGNY G+ C Y +PLQG
Sbjct: 183 LALEAARQGIVLLENKGNALPLSPTRHRTVGVIGPNSDVTVTMIGNYAGVACGYTTPLQG 362
Query: 451 LTALVPTSYVPGCPDVHCASAQL-DDATKIAASSDATVIVVGASLAIEAESLDRVNIVLP 509
+ V T + GC DV C +L +A +A SDATV+VVG +IEAE DRV ++LP
Sbjct: 363 IARYVKTIHQAGCRDVSCRGTELFGNAEIVARQSDATVVVVGLDQSIEAEFRDRVGLLLP 542
Query: 510 GQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVI 569
G QQ LVS VA A+RGPV+LVIMSGG +DV FAK + KI++ILWVGYPG+AGG AIADVI
Sbjct: 543 GHQQELVSRVARAARGPVVLVIMSGGPVDVQFAKDDPKISAILWVGYPGQAGGTAIADVI 722
Query: 570 FGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYKGETVFSFGDGIGY 629
FG NP GRLP TWYP+SYV +PMTNM+MR +P++GYPGRTYRFYKG VF FG G+ Y
Sbjct: 723 FGRTNPGGRLPNTWYPQSYVAKLPMTNMDMRPNPASGYPGRTYRFYKGPVVFPFGHGLSY 902
Query: 630 SNVEHKIVKAPQLVFVPLAEDHVCRSS--ECKSLDVADEHCQNMAFDIHLRVKNMGKMSS 687
S H + AP+ V VP+A R+S K++ V+ C + H+ VKN G M
Sbjct: 903 SRFTHTLAVAPEQVSVPIASLQAFRNSTMSSKAVKVSHAKCGALKMGFHVDVKNEGSMDG 1082
Query: 688 SHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPL 747
+HT+L+F TPP + K L+ K ++ S+ +V V VCK LSVVDE G RR+P+
Sbjct: 1083THTLLIFSTPPPGKWSASKQLVSFHKTYVPAGSKQRVNVGVHVCKHLSVVDENGIRRIPM 1262
Query: 748 GQHLLHVGNLKYPLSVR 764
G H LH+G++K+ +SV+
Sbjct: 1263GDHELHIGDVKHTISVQ 1313
>TC17591 similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (24%)
Length = 582
Score = 388 bits (997), Expect = e-108
Identities = 192/193 (99%), Positives = 192/193 (99%)
Frame = +3
Query: 277 LKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE 336
LKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE
Sbjct: 3 LKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE 182
Query: 337 GAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAA 396
GAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAA
Sbjct: 183 GAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAA 362
Query: 397 RQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGNYEGIPCKYISPLQGLTALVP 456
RQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGN EGIPCKYISPLQGLTALVP
Sbjct: 363 RQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGN*EGIPCKYISPLQGLTALVP 542
Query: 457 TSYVPGCPDVHCA 469
TSYVPGCPDVHCA
Sbjct: 543 TSYVPGCPDVHCA 581
>TC19191 similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragment),
partial (31%)
Length = 770
Score = 352 bits (904), Expect = 1e-97
Identities = 163/208 (78%), Positives = 184/208 (88%)
Frame = +2
Query: 558 GEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYKG 617
GEAGGAAIADVIFGF+NPSGRLPMTWYP+SYV+ VPMTNMNMR DP+TGYPGRTYRFYKG
Sbjct: 2 GEAGGAAIADVIFGFHNPSGRLPMTWYPQSYVDKVPMTNMNMRPDPTTGYPGRTYRFYKG 181
Query: 618 ETVFSFGDGIGYSNVEHKIVKAPQLVFVPLAEDHVCRSSECKSLDVADEHCQNMAFDIHL 677
ETVFSFGDG+ YS +EHK+VKAPQLV +PLAEDHVC S +CKSLDVA EHC+N+ F+IHL
Sbjct: 182 ETVFSFGDGLSYSTIEHKLVKAPQLVSIPLAEDHVCHSLKCKSLDVAGEHCENLEFEIHL 361
Query: 678 RVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVV 737
+KN G+MSS HTV LF +PPAVHNAP KHL+G EKVHL G +EA V+F VDVCKDLSVV
Sbjct: 362 SIKNKGRMSSDHTVFLFSSPPAVHNAPHKHLVGFEKVHLEGNTEALVKFKVDVCKDLSVV 541
Query: 738 DELGNRRVPLGQHLLHVGNLKYPLSVRI 765
DELGNR+V LGQHLLHVG+LK+ L V I
Sbjct: 542 DELGNRKVALGQHLLHVGDLKHALGVMI 625
>TC19239 homologue to UP|Q9ZU04 (Q9ZU04) Beta-glucosidase (Fragment) ,
partial (69%)
Length = 453
Score = 307 bits (787), Expect = 4e-84
Identities = 150/151 (99%), Positives = 151/151 (99%)
Frame = +1
Query: 497 EAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGY 556
+AESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGY
Sbjct: 1 QAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGY 180
Query: 557 PGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYK 616
PGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYK
Sbjct: 181 PGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRTYRFYK 360
Query: 617 GETVFSFGDGIGYSNVEHKIVKAPQLVFVPL 647
GETVFSFGDGIGYSNVEHKIVKAPQLVFVPL
Sbjct: 361 GETVFSFGDGIGYSNVEHKIVKAPQLVFVPL 453
>BP041585
Length = 503
Score = 268 bits (686), Expect = 2e-72
Identities = 122/167 (73%), Positives = 147/167 (87%), Gaps = 3/167 (1%)
Frame = +1
Query: 85 PRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASFNSTLFQAIGKVVSTEARA 144
P YEWWSEALHGVSN+GPGTRF+ VPGATSFP IL+AASFN+TL+ +G+VVSTEARA
Sbjct: 1 PSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNATLWYNMGQVVSTEARA 180
Query: 145 MYNVGLAGLTYWSPNINIFRDPRWGRGQETPGEDPLLSSKYAAGYVKGLQQTDDGDS--- 201
MYNV LAGLT+WSPN+N+FRDPRWGRGQETPGEDPL+ S+YA YV+GLQ +D +S
Sbjct: 181 MYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQDVEDEESIKA 360
Query: 202 NKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLDDTFQPPFKS 248
++LKV++CCKHYTAYDVDNWKGV R+ F+A V++QDL+DT+QPPFKS
Sbjct: 361 DRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVTKQDLEDTYQPPFKS 501
>BP033690
Length = 546
Score = 226 bits (577), Expect = 8e-60
Identities = 112/142 (78%), Positives = 122/142 (85%)
Frame = +2
Query: 17 FFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQEKIGNLGDSA 76
F C V GQT+P FACDV KN + +G+GFC++SLPV RVADLV RLTLQEKIGNL A
Sbjct: 119 FSCGLVWGQTTPNFACDVAKNPALSGYGFCNRSLPVNARVADLVGRLTLQEKIGNLVSGA 298
Query: 77 VDVGRLGIPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASFNSTLFQAIGK 136
V RLGIP+YEWWSEALHGVSNIGPGTRFS+VVP A SFPMPILTAASFN++LFQAIGK
Sbjct: 299 AIVSRLGIPKYEWWSEALHGVSNIGPGTRFSNVVPRAASFPMPILTAASFNTSLFQAIGK 478
Query: 137 VVSTEARAMYNVGLAGLTYWSP 158
VVSTEARAMYNVGLAGLTY SP
Sbjct: 479 VVSTEARAMYNVGLAGLTYGSP 544
>TC15933 weakly similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragment),
partial (14%)
Length = 453
Score = 180 bits (456), Expect = 9e-46
Identities = 88/91 (96%), Positives = 90/91 (98%)
Frame = +1
Query: 675 IHLRVKNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDL 734
IHLRVKNMGKM+SSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDL
Sbjct: 1 IHLRVKNMGKMTSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDL 180
Query: 735 SVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
SVVDEL +RRVPLGQHLLHVGNLKYPLSVRI
Sbjct: 181 SVVDELAHRRVPLGQHLLHVGNLKYPLSVRI 273
>CN825311
Length = 522
Score = 177 bits (449), Expect = 6e-45
Identities = 85/163 (52%), Positives = 111/163 (67%), Gaps = 3/163 (1%)
Frame = +2
Query: 183 SKYAAGYVKGL--QQTDDGDS-NKLKVAACCKHYTAYDVDNWKGVQRYTFNAVVSQQDLD 239
S Y G G +DDGD + L V+ACCKH+TAYD++ W RY FNAVVSQQDL+
Sbjct: 32 SDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYNFNAVVSQQDLE 211
Query: 240 DTFQPPFKSCVIDGNVASVMCSYNQVNGKPTCADPDLLKGVIRGQWKLNGYIVSDCDSVE 299
DT+QPPF+ CV G + +MCSYN+VNG P CA DLL GV R W GYI SDCD+V
Sbjct: 212 DTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLL-GVARNNWGFKGYITSDCDAVA 388
Query: 300 VLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQG 342
+F+ Q Y K+ E+A A+ + AG D++CG+Y+ ++T AV+QG
Sbjct: 389 TVFEYQVYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQG 517
>AV770828
Length = 527
Score = 174 bits (440), Expect = 6e-44
Identities = 93/168 (55%), Positives = 120/168 (71%), Gaps = 1/168 (0%)
Frame = +1
Query: 377 GLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGN 436
G GP+DVCT +QELA EAARQGIVLLKNS SLPLS + ++LA+IGPN+NAT MIGN
Sbjct: 16 GTGPRDVCTQPHQELALEAARQGIVLLKNSGPSLPLSTRRHRTLAIIGPNSNATVTMIGN 195
Query: 437 YEGIPCKYISPLQGLTALVPTSYVPGCPDV-HCASAQLDDATKIAASSDATVIVVGASLA 495
Y GI +Y SP +G+ T + GC +V Q A A +DATV+V+G +
Sbjct: 196 YAGIGYRYTSP*EGIGKYAKTIHELGCANVAGTDDKQFGSALNAARQADATVLVMGLDQS 375
Query: 496 IEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAK 543
IEAE++DR ++LPG+QQ LVS+VA AS+GP ILV+MSGG +D++FAK
Sbjct: 376 IEAETVDRAGLLLPGRQQDLVSKVAAASKGPTILVLMSGGPVDITFAK 519
>TC12358 weakly similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (6%)
Length = 395
Score = 149 bits (375), Expect = 2e-36
Identities = 71/71 (100%), Positives = 71/71 (100%)
Frame = +1
Query: 1 MACLLQNRVSVLVCFSFFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLV 60
MACLLQNRVSVLVCFSFFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLV
Sbjct: 181 MACLLQNRVSVLVCFSFFCATVCGQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLV 360
Query: 61 KRLTLQEKIGN 71
KRLTLQEKIGN
Sbjct: 361 KRLTLQEKIGN 393
>TC18444 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial (12%)
Length = 387
Score = 131 bits (329), Expect = 5e-31
Identities = 57/91 (62%), Positives = 71/91 (77%)
Frame = +2
Query: 547 KITSILWVGYPGEAGGAAIADVIFGFYNPSGRLPMTWYPESYVENVPMTNMNMRADPSTG 606
KI ILW GYPGE GG A+A VIFG +NP GRLP+TWYP+ Y++ VPMT+M MRADP+TG
Sbjct: 2 KIGGILWAGYPGELGGIALAQVIFGDHNPGGRLPITWYPKDYIK-VPMTDMRMRADPATG 178
Query: 607 YPGRTYRFYKGETVFSFGDGIGYSNVEHKIV 637
YPGRTYRFYKG ++ FG G+ YS ++ +
Sbjct: 179 YPGRTYRFYKGPKLYEFGYGLSYSKYSYEFI 271
>AV775420
Length = 457
Score = 130 bits (328), Expect = 6e-31
Identities = 70/143 (48%), Positives = 95/143 (65%), Gaps = 1/143 (0%)
Frame = +1
Query: 309 KTPEEAAAKSILAGLDLDCGSYLGKYTEGAVKQGLLDEASINNAISNNFATLMRLGFFDG 368
K A + AG+D++CG YL K+ + AV Q + + I+ A+ N F+ +RLG FDG
Sbjct: 1 KNSRGCVADVLRAGMDVECGDYLTKHGKSAV*QKKVPISQIDRALHNLFSIRIRLGLFDG 180
Query: 369 NPSTLPYGGLGPKDVCTSENQELAREAARQGIVLLKNSPGSLPLSAKSIKSLAVIGPNAN 428
NPS L YG +GP VC+ +N +LA EAAR GIVLLK + LPL K+ S+AVIGPNAN
Sbjct: 181 NPSKLTYGSIGPNQVCSKQNLKLALEAARNGIVLLKKTASILPL-PKTNPSMAVIGPNAN 357
Query: 429 ATRV-MIGNYEGIPCKYISPLQG 450
A+ + ++GNY G PCK ++ LQG
Sbjct: 358 ASSLAVLGNYYGRPCKLVTLLQG 426
>TC13306 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (12%)
Length = 435
Score = 119 bits (297), Expect = 2e-27
Identities = 59/103 (57%), Positives = 72/103 (69%)
Frame = +2
Query: 24 GQTSPPFACDVTKNASFAGFGFCDKSLPVADRVADLVKRLTLQEKIGNLGDSAVDVGRLG 83
G+ PFACD K + GF FC ++P+ RV DL+ RLTL EKI + ++A++V RLG
Sbjct: 128 GEARVPFACDPQKGLT-RGFKFCRTNVPIHVRVQDLIGRLTLAEKIRLVVNNAIEVPRLG 304
Query: 84 IPRYEWWSEALHGVSNIGPGTRFSSVVPGATSFPMPILTAASF 126
I YEWWSEALHGVSN+GPGT+F P ATSFP I TAASF
Sbjct: 305 IQGYEWWSEALHGVSNVGPGTKFGGAFPAATSFPQVITTAASF 433
>TC19733 weakly similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, partial
(15%)
Length = 598
Score = 97.4 bits (241), Expect = 7e-21
Identities = 45/108 (41%), Positives = 71/108 (65%), Gaps = 2/108 (1%)
Frame = +2
Query: 659 KSLDVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTPP--AVHNAPQKHLLGIEKVHL 716
K++ V C ++ +H+ VKN+G +HT+L+F PP H A QK L+ EKVH+
Sbjct: 62 KAIRVTHARCGKLSIALHVNVKNVGSRDGTHTLLVFSAPPDGGAHWAVQKQLVAFEKVHV 241
Query: 717 AGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVR 764
KS+ +V+ + VCK LSVVD G RR+P+G+H LH+G++K+ +S++
Sbjct: 242 PAKSQQRVQINIHVCKLLSVVDRSGIRRIPMGEHSLHIGDVKHSVSLQ 385
>BP075556
Length = 469
Score = 74.7 bits (182), Expect = 5e-14
Identities = 40/77 (51%), Positives = 49/77 (62%), Gaps = 1/77 (1%)
Frame = -3
Query: 680 KNMGKMSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSEAQV-RFMVDVCKDLSVVD 738
+N G+MSS HT+ LF +PP V P L GI + L K + +F VDVCKDLSVVD
Sbjct: 443 QNQGRMSSDHTLFLFSSPPPVPTPPSSTLTGI*ESSLGRKYRTPLAKFKVDVCKDLSVVD 264
Query: 739 ELGNRRVPLGQHLLHVG 755
ELG+ +V LGQHL G
Sbjct: 263 ELGHSQVALGQHLASCG 213
Score = 39.3 bits (90), Expect = 0.002
Identities = 36/103 (34%), Positives = 50/103 (47%), Gaps = 8/103 (7%)
Frame = -1
Query: 671 MAFDIHLRVKNMGK-----MSSSHTVLLFFTPPAVHNAPQKHLLGIEKVHLAGKSE---A 722
+ F+IHL ++ G+ +SS +LL P +H P HLLG EKVHL G +E
Sbjct: 469 LEFEIHLSLRTRGE*AVITLSSCSLLLL---PCPLH--PHPHLLGFEKVHLEGNTERPWL 305
Query: 723 QVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
R+M K + L ++ LHVG LK+ L V I
Sbjct: 304 SSRWM--FVKI*VWLMNLATAKLH*DSISLHVGELKHALGVMI 182
>AV427695
Length = 202
Score = 74.3 bits (181), Expect = 7e-14
Identities = 38/64 (59%), Positives = 49/64 (76%)
Frame = +2
Query: 476 ATKIAASSDATVIVVGASLAIEAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGG 535
A A +DATV+V+G +IEAE+ DRV ++LPG QQ LVS+VA ASRGP ILV+MSGG
Sbjct: 11 ALDAARHADATVLVMGLDQSIEAETRDRVGLLLPGHQQDLVSKVAAASRGPTILVLMSGG 190
Query: 536 GMDV 539
+D+
Sbjct: 191 PLDI 202
>AV415013
Length = 154
Score = 68.9 bits (167), Expect = 3e-12
Identities = 32/50 (64%), Positives = 37/50 (74%)
Frame = +2
Query: 531 IMSGGGMDVSFAKANDKITSILWVGYPGEAGGAAIADVIFGFYNPSGRLP 580
IM+ G +D+SF K+ I ILWVGYPG+AGG AIA VIFG YNP GR P
Sbjct: 2 IMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGGRSP 151
>TC15428
Length = 629
Score = 43.5 bits (101), Expect(2) = 6e-11
Identities = 23/61 (37%), Positives = 33/61 (53%), Gaps = 1/61 (1%)
Frame = +1
Query: 662 DVADEHCQNMAFDIHLRVKNMGKMSSSHTVLLFFTPPAVHNA-PQKHLLGIEKVHLAGKS 720
++ +E CQ+M+ + L VKN G M+ H VLLF P N P K L+G V L +
Sbjct: 115 ELGEETCQSMSVSVTLGVKNDGSMAGKHPVLLFMRPGKQRNGNPLKPLVGFPSVKLDARG 294
Query: 721 E 721
+
Sbjct: 295 K 297
Score = 40.8 bits (94), Expect(2) = 6e-11
Identities = 16/43 (37%), Positives = 30/43 (69%)
Frame = +2
Query: 723 QVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVRI 765
+V F + C+ LS+ +E G + +P G +LLHVG+ +YP+++ +
Sbjct: 302 EVGFELSPCEHLSIANEGGVKVIPEGSYLLHVGDQEYPMNISV 430
>AV780323
Length = 544
Score = 61.2 bits (147), Expect = 6e-10
Identities = 26/60 (43%), Positives = 43/60 (71%)
Frame = -3
Query: 705 QKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSVVDELGNRRVPLGQHLLHVGNLKYPLSVR 764
+K L+ EKVH+ K++ +VR + VCK LSVVD G RR+P+G+H +G++++ LS++
Sbjct: 542 RKQLVAFEKVHIPAKAQRRVRVNIHVCKLLSVVDRSGIRRIPMGEHRFQIGDIEHSLSLQ 363
>BP028363
Length = 508
Score = 59.3 bits (142), Expect = 2e-09
Identities = 32/80 (40%), Positives = 46/80 (57%), Gaps = 3/80 (3%)
Frame = -3
Query: 680 KNMGKMSSSHTVLLFFTPPA---VHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKDLSV 736
KN G + SH VL+F+ PP V AP K L+G E+ H+ V +D+C+ LS
Sbjct: 506 KNTGPYNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLSN 327
Query: 737 VDELGNRRVPLGQHLLHVGN 756
VD G R++ +G+H L VG+
Sbjct: 326 VDSDGKRKLIIGKHTLLVGS 267
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,052,356
Number of Sequences: 28460
Number of extensions: 176192
Number of successful extensions: 814
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 798
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 803
length of query: 765
length of database: 4,897,600
effective HSP length: 97
effective length of query: 668
effective length of database: 2,136,980
effective search space: 1427502640
effective search space used: 1427502640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0314.7


