

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0314.22
(324 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
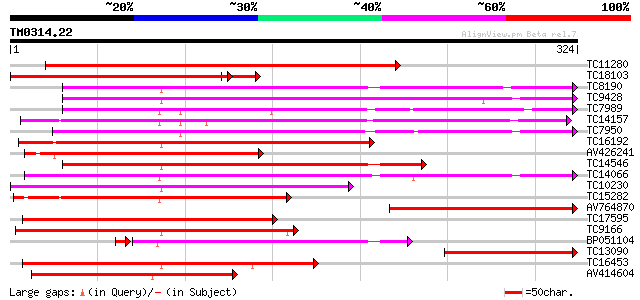
Score E
Sequences producing significant alignments: (bits) Value
TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), p... 377 e-105
TC18103 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 ... 252 1e-72
TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 p... 251 1e-67
TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), pa... 238 8e-64
TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , par... 229 4e-61
TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , p... 218 1e-57
TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), part... 208 9e-55
TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%) 197 3e-51
AV426241 195 1e-50
TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxida... 186 4e-48
TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 ,... 179 4e-46
TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precurso... 169 8e-43
TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fra... 165 1e-41
AV764870 161 1e-40
TC17595 similar to UP|Q9XFI7 (Q9XFI7) Peroxidase (Fragment), par... 155 9e-39
TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%) 139 6e-34
BP051104 136 4e-33
TC13090 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 ... 136 5e-33
TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, part... 135 7e-33
AV414604 129 9e-31
>TC11280 similar to UP|Q9XFL5 (Q9XFL5) Peroxidase 4 (Fragment), partial
(68%)
Length = 614
Score = 377 bits (969), Expect = e-105
Identities = 192/204 (94%), Positives = 193/204 (94%), Gaps = 1/204 (0%)
Frame = +3
Query: 21 TVHGQG-SRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVL 79
TVHGQG SRVGFY TCPRAESIVRS VESHV SD TLAAGLLRMHFHDCFVQGCDASVL
Sbjct: 3 TVHGQGGSRVGFYLGTCPRAESIVRSTVESHVNSDPTLAAGLLRMHFHDCFVQGCDASVL 182
Query: 80 IAGAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQ 139
IAGAGTERTA PNL LRG+EVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQ
Sbjct: 183 IAGAGTERTAIPNLSLRGFEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQ 362
Query: 140 VPTGRRDGRVSQASDVNNLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFF 199
VPTGRRDGRVSQASDVNNLPAPFDSVDVQKQKF AKGLNTQDLVTLVGGHTIGTTACQFF
Sbjct: 363 VPTGRRDGRVSQASDVNNLPAPFDSVDVQKQKFTAKGLNTQDLVTLVGGHTIGTTACQFF 542
Query: 200 SNRLYNFTSNGPDSSIDASFLPQL 223
SNRLYNFTSNGPD SIDASFL QL
Sbjct: 543 SNRLYNFTSNGPDPSIDASFLLQL 614
>TC18103 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 precursor
(PNPC2) , partial (37%)
Length = 546
Score = 252 bits (643), Expect(2) = 1e-72
Identities = 127/127 (100%), Positives = 127/127 (100%)
Frame = +1
Query: 1 MERSLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAG 60
MERSLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAG
Sbjct: 115 MERSLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAG 294
Query: 61 LLRMHFHDCFVQGCDASVLIAGAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCAD 120
LLRMHFHDCFVQGCDASVLIAGAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCAD
Sbjct: 295 LLRMHFHDCFVQGCDASVLIAGAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCAD 474
Query: 121 ILALAAR 127
ILALAAR
Sbjct: 475 ILALAAR 495
Score = 38.1 bits (87), Expect(2) = 1e-72
Identities = 18/22 (81%), Positives = 18/22 (81%)
Frame = +2
Query: 122 LALAARDSVVLSGGLSWQVPTG 143
L L DSVVLSGGLSWQVPTG
Sbjct: 479 LPLLLDDSVVLSGGLSWQVPTG 544
>TC8190 similar to UP|PER1_ARAHY (P22195) Cationic peroxidase 1 precursor
(PNPC1) , partial (96%)
Length = 1202
Score = 251 bits (641), Expect = 1e-67
Identities = 136/299 (45%), Positives = 178/299 (59%), Gaps = 5/299 (1%)
Frame = +1
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT---ER 87
FY +TCP + +++ V V + + A LLR+HFHDCFVQGCDAS+L+ + E+
Sbjct: 154 FYAKTCPLVLATIKTQVNLAVAKEARMGASLLRLHFHDCFVQGCDASILLDDTSSFTGEK 333
Query: 88 TAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
TA PN +RGY+VID K+KVE+ CPGVVSCADI+A+AARDSVV GG SW VP GRRD
Sbjct: 334 TAGPNANSVRGYDVIDTIKSKVESLCPGVVSCADIVAVAARDSVVALGGFSWAVPLGRRD 513
Query: 147 GRVSQASDVNN-LPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
+ S N+ LP P ++D F+ KG T+++V L G HTIG C FF R+Y
Sbjct: 514 STTASLSSANSELPGPSSNLDGLNTAFSNKGFTTREMVALSGSHTIGQARCLFFRTRIYY 693
Query: 206 FTSNGPDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLSD 265
++ ID++F LQ CP N G SN LDTTS FD YY NL++ +G+ SD
Sbjct: 694 ------ETHIDSTFAKNLQGNCPFNGGDSNLSPLDTTSPTTFDNGYYRNLQSQKGLFHSD 855
Query: 266 QALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSAFN 324
Q L+N ST + V Y+ F +F +MVKM N+ TGS +IR C N
Sbjct: 856 QVLFNGGSTDSQVNSYV----TNPASFKTDFANAMVKMGNLSPLTGSSGQIRTHCRKTN 1020
>TC9428 similar to UP|Q9XFL3 (Q9XFL3) Peroxidase 1 (Fragment), partial
(97%)
Length = 1354
Score = 238 bits (608), Expect = 8e-64
Identities = 132/302 (43%), Positives = 177/302 (57%), Gaps = 8/302 (2%)
Frame = +2
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT----E 86
FY+ TCP SIVR V + KSD + L+R+HFHDCFVQGCDAS+L+ T +
Sbjct: 128 FYKDTCPNVHSIVREVVRNVSKSDPRMLGSLIRLHFHDCFVQGCDASILLNDTSTIVSEQ 307
Query: 87 RTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRRD 146
P N +RG +V++ K VE ACP VSCADILALAA S VL+ G+ W+VP GRRD
Sbjct: 308 GAFPNNNSIRGLDVVNQIKTAVENACPNTVSCADILALAAEISSVLANGVDWKVPLGRRD 487
Query: 147 GRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLYN 205
+ S N NLP P ++ FA + LN DLV L G HTIG CQFF NRLYN
Sbjct: 488 SLTANRSLANQNLPGPNFNLSQLNASFAVQNLNITDLVALSGAHTIGRGQCQFFVNRLYN 667
Query: 206 FTSNG-PDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLS 264
F++ G PD +++ ++L L+++CP + LD T+ + D+ Y++NL+ G+G+ S
Sbjct: 668 FSNTGNPDPTLNTTYLQTLRSICPHGGPGTTLAHLDPTTPDTLDSQYFSNLQGGKGLFQS 847
Query: 265 DQALW--NDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSA 322
D L+ + A+T V + L F F SM+KMS I + TGS EIRK C+
Sbjct: 848 DPVLFSTSGAATVAIVNSFSSNPTL----FFENFKASMIKMSRIGVLTGSQGEIRKPCNF 1015
Query: 323 FN 324
N
Sbjct: 1016VN 1021
>TC7989 similar to UP|Q43854 (Q43854) Peroxidase precursor , partial (85%)
Length = 1352
Score = 229 bits (585), Expect = 4e-61
Identities = 131/301 (43%), Positives = 178/301 (58%), Gaps = 7/301 (2%)
Frame = +3
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAG---TER 87
FY +CP+ ESI+R ++ D AAGLLR+HFHDCFVQGCDASVL+ G+ +E+
Sbjct: 150 FYDSSCPKLESIIRKELKKVFDKDIAQAAGLLRLHFHDCFVQGCDASVLLDGSASGPSEK 329
Query: 88 TAPPNLGLR--GYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRR 145
APPNL LR +++I+D ++++E C VVSCADI A+AARD+V LSGG +++P GRR
Sbjct: 330 DAPPNLTLRAQAFKIIEDLRSQIEKKCGRVVSCADITAIAARDAVFLSGGPDYELPLGRR 509
Query: 146 DGR--VSQASDVNNLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRL 203
DG ++ ++NLPAP + A K L+ D+V+L GGHTIG + C F++RL
Sbjct: 510 DGLNFATRNVTLDNLPAPQSNTTTILNSLATKNLDPTDVVSLSGGHTIGISHCSSFTDRL 689
Query: 204 YNFTSNGPDSSIDASFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILL 263
Y D +D +F L+ CP S N LD S N FD YY +L N +G+
Sbjct: 690 Y----PSKDPVMDQTFEKNLKLTCPA-SNTDNTTVLDLRSPNTFDNKYYVDLMNRQGLFF 854
Query: 264 SDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSAF 323
SDQ L+ D TK V + + L KF V +M+KM + + TGS EIR CS
Sbjct: 855 SDQDLYTDKRTKDIVTSFAVNQSLFFEKFVV----AMLKMGQLNVLTGSQGEIRANCSVR 1022
Query: 324 N 324
N
Sbjct: 1023N 1025
>TC14157 similar to UP|O24080 (O24080) Peroxidase2 precursor , partial
(96%)
Length = 1390
Score = 218 bits (554), Expect = 1e-57
Identities = 129/323 (39%), Positives = 180/323 (54%), Gaps = 8/323 (2%)
Frame = +1
Query: 7 RIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHF 66
R+ L L+L T Q S Y TCP +SIV+ V+ + LR+ F
Sbjct: 70 RVLVLSLSLTLCFYTCFAQLSP-NHYASTCPNLQSIVKGVVQKKFQQTFVTVPATLRLFF 246
Query: 67 HDCFVQGCDASVLIAGAG---TERTAPPNLGLR--GYEVIDDAKAKVEAA--CPGVVSCA 119
HDCFVQGCDASV++A +G E+ P NL L G++ + AKA V+A C VSCA
Sbjct: 247 HDCFVQGCDASVMVASSGNNKAEKDHPDNLSLAGDGFDTVIKAKAAVDAVPQCRNKVSCA 426
Query: 120 DILALAARDSVVLSGGLSWQVPTGRRDGRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLN 178
DILALA RD VVL+GG S+ V GR DG VS+ASDVN LP P +++ FA++GL
Sbjct: 427 DILALATRDVVVLAGGPSYTVELGRFDGLVSRASDVNGRLPEPNFNLNQLNSLFASQGLT 606
Query: 179 TQDLVTLVGGHTIGTTACQFFSNRLYNFTSNGPDSSIDASFLPQLQALCPQNSGVSNRVA 238
D++ L G HT+G + C FSNR+Y S D +++ ++ QLQ +CP+N +
Sbjct: 607 QTDMIALSGAHTLGFSHCNRFSNRIY---STPVDPTLNRNYATQLQQMCPKNVNPQIAIN 777
Query: 239 LDTTSQNRFDTSYYANLRNGRGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGK 298
+D T+ FD YY NL+ G+G+ SDQ L+ D +K V + FN F
Sbjct: 778 MDPTTPRTFDNIYYKNLQQGKGLFTSDQILFTDQRSKATVNSFASNSN----TFNANFAA 945
Query: 299 SMVKMSNIELKTGSDSEIRKICS 321
+M+K+ + +KT + +IR CS
Sbjct: 946 AMIKLGRVGVKTARNGKIRTDCS 1014
>TC7950 similar to UP|Q9XFI8 (Q9XFI8) Peroxidase (Fragment), partial (86%)
Length = 1286
Score = 208 bits (530), Expect = 9e-55
Identities = 119/303 (39%), Positives = 170/303 (55%), Gaps = 3/303 (0%)
Frame = +1
Query: 25 QGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAG 84
+G FY +TCP+ E++VR+ ++ +K D A GLLR+ FHDCFVQGCD SVL+ G+
Sbjct: 100 EGLSFSFYSKTCPKLETVVRNHLKKVLKKDNGQAPGLLRIFFHDCFVQGCDGSVLLDGSP 279
Query: 85 TERTAPPNLGLR--GYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPT 142
ER P N+G+R + I+D +A V C +VSCADI LA+RD+V L+GG + VP
Sbjct: 280 GERDQPANIGIRPEALQTIEDIRALVHKQCGKIVSCADITILASRDAVFLTGGPDYAVPL 459
Query: 143 GRRDGRVSQASDVNNLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNR 202
GRRDG LP+P ++ + FA + + D+V L G HT G C F NR
Sbjct: 460 GRRDGVSFSTVGTQKLPSPINNTTATLKAFADRNFDATDVVALSGAHTFGRAHCGTFFNR 639
Query: 203 LYNFTSNGPDSSIDASFLPQLQALCP-QNSGVSNRVALDTTSQNRFDTSYYANLRNGRGI 261
L + D ++D + L A CP QNS +N LD + N FD YY +L N +G+
Sbjct: 640 L-----SPLDPNMDKTLAKNLTATCPAQNS--TNTANLDIRTPNVFDNKYYLDLMNRQGV 798
Query: 262 LLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICS 321
SDQ L +D TK V + + L F +F +++K+S +++ TG+ EIR C+
Sbjct: 799 FTSDQDLLSDKRTKGLVNAFAVNQTL----FFEKFVDAVIKLSQLDVLTGNQGEIRGRCN 966
Query: 322 AFN 324
N
Sbjct: 967 VVN 975
>TC16192 similar to UP|Q9SSZ9 (Q9SSZ9) Peroxidase 1, partial (49%)
Length = 645
Score = 197 bits (500), Expect = 3e-51
Identities = 106/208 (50%), Positives = 140/208 (66%), Gaps = 5/208 (2%)
Frame = +2
Query: 6 FRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMH 65
F A ++L + + + + VG+Y +C AE IV+ V V + +AAGL+RMH
Sbjct: 23 FNCAIIVLVIFLLNQNAYSE-LEVGYYSYSCGMAEFIVKDEVRRSVTKNPGIAAGLVRMH 199
Query: 66 FHDCFVQGCDASVLIAGAGT---ERTAPPNL-GLRGYEVIDDAKAKVEAACPGVVSCADI 121
FHDCF++GCDASVL+ + E+ +P N LRG+EVID AKAK+EA C GVVSCADI
Sbjct: 200 FHDCFIRGCDASVLLDSTPSNTAEKDSPANKPSLRGFEVIDSAKAKLEAVCKGVVSCADI 379
Query: 122 LALAARDSVVLSGGLSWQVPTGRRDGRVSQASDV-NNLPAPFDSVDVQKQKFAAKGLNTQ 180
+A AARDSV L+GG+ + VP GRRDGR+S ASD +LP P +V+ Q FA KGL +
Sbjct: 380 IAFAARDSVELAGGVGYDVPAGRRDGRISLASDTRTDLPPPTFNVNQLTQLFAKKGLTQE 559
Query: 181 DLVTLVGGHTIGTTACQFFSNRLYNFTS 208
++VTL G HTIG + C FS+RLYNF+S
Sbjct: 560 EMVTLSGAHTIGRSHCAAFSSRLYNFSS 643
>AV426241
Length = 429
Score = 195 bits (495), Expect = 1e-50
Identities = 100/140 (71%), Positives = 115/140 (81%), Gaps = 3/140 (2%)
Frame = +3
Query: 9 AFLLLALASIVNTVHG---QGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMH 65
+FLL+ L IV T+ G+ VGFY ++CP ESIV+S V SHVK+D AAGLLR+H
Sbjct: 15 SFLLVFL--IVLTLQAFAVHGTSVGFYSKSCPSIESIVKSTVASHVKTDFEYAAGLLRLH 188
Query: 66 FHDCFVQGCDASVLIAGAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALA 125
F DCFV+GCDAS+LIAG GTE+ APPN L+GYEVID+AKAK+EA CPGVVSCADILALA
Sbjct: 189 FRDCFVRGCDASILIAGNGTEKQAPPNRSLKGYEVIDEAKAKLEAQCPGVVSCADILALA 368
Query: 126 ARDSVVLSGGLSWQVPTGRR 145
ARDSVVLSGGLSWQVPTGRR
Sbjct: 369 ARDSVVLSGGLSWQVPTGRR 428
>TC14546 similar to UP|Q9XGV6 (Q9XGV6) Bacterial-induced peroxidase
precursor , partial (66%)
Length = 738
Score = 186 bits (473), Expect = 4e-48
Identities = 97/214 (45%), Positives = 131/214 (60%), Gaps = 6/214 (2%)
Frame = +1
Query: 31 FYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQGCDASVLIAGAGT----E 86
FY R+CP+ ++IVR+A+ +K + + A +LR+ FHDCFV GCD S+L+ GT E
Sbjct: 112 FYDRSCPKLQTIVRNAMVQTIKKEARMGASILRLFFHDCFVNGCDGSILLDDIGTTFVGE 291
Query: 87 RTAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCADILALAARDSVVLSGGLSWQVPTGRR 145
+ A PN RG+EVID K VEA+C VSCADILALA RD + L GG +WQVP GRR
Sbjct: 292 KNAAPNKNSARGFEVIDTIKTNVEASCNNTVSCADILALATRDGINLLGGPTWQVPLGRR 471
Query: 146 DGRVSQASDVN-NLPAPFDSVDVQKQKFAAKGLNTQDLVTLVGGHTIGTTACQFFSNRLY 204
D R + S N +P+P + F+AKGL+ +DL L GGHTIG CQFF +R+
Sbjct: 472 DARTASQSKANTEIPSPSSDLSTLISMFSAKGLSARDLTVLSGGHTIGQAECQFFRSRVN 651
Query: 205 NFTSNGPDSSIDASFLPQLQALCPQNSGVSNRVA 238
N +++IDA+F + CP + G +A
Sbjct: 652 N------ETNIDAAFAASRKTNCPASGGGDTNLA 735
>TC14066 homologue to UP|O64970 (O64970) Cationic peroxidase 2 , partial
(92%)
Length = 1436
Score = 179 bits (455), Expect = 4e-46
Identities = 113/323 (34%), Positives = 165/323 (50%), Gaps = 7/323 (2%)
Frame = +2
Query: 9 AFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHD 68
A LL+ +S V G + FY+ TCP+AE I+ V+ K + A LR FHD
Sbjct: 98 ALSLLSPSSAQEDVQDTGLAMNFYKETCPQAEDIITEQVKLLYKRHKNTAFSWLRNIFHD 277
Query: 69 CFVQGCDASVLIAGAG---TERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALA 125
C VQ CDAS+L+ +E+ + GLR + I+ K +E CPGVVSC+DIL L+
Sbjct: 278 CAVQSCDASLLLDSTRKTLSEKETDRSFGLRNFRYIETIKEALERECPGVVSCSDILVLS 457
Query: 126 ARDSVVLSGGLSWQVPTGRRDGRVSQASDVNN-LPAPFDSVDVQKQKFAAKGLNTQDLVT 184
ARD + GG + TGRRDGR S+A V LP +S+ KF A G++T +V
Sbjct: 458 ARDGIAALGGPHIPLRTGRRDGRRSRADVVEQFLPDHNESISAVLDKFGAMGIDTPGVVA 637
Query: 185 LVGGHTIGTTACQFFSNRLYNFTSNGPDSSIDASFLPQLQALCPQ---NSGVSNRVALDT 241
L+G H++G T C +RLY D +++ +P + CP + V D
Sbjct: 638 LLGAHSVGRTHCVKLVHRLYPEV----DPALNPDHIPHMLKKCPDAIPDPKAVQYVRNDR 805
Query: 242 TSQNRFDTSYYANLRNGRGILLSDQALWNDASTKTFVRRYLGLRGLLGLKFNVEFGKSMV 301
+ D +YY N+ + +G+LL D L ND TK +V++ + F EF +++
Sbjct: 806 GTPMILDNNYYRNILDNKGLLLVDHQLANDKRTKPYVKKMAKSQDY----FFKEFSRAIT 973
Query: 302 KMSNIELKTGSDSEIRKICSAFN 324
+S TG+ EIRK C+ N
Sbjct: 974 LLSENNPLTGTKGEIRKQCNVAN 1042
>TC10230 similar to UP|PE10_ARATH (Q9FX85) Peroxidase 10 precursor (Atperox
P10) (ATP5a) , partial (42%)
Length = 753
Score = 169 bits (427), Expect = 8e-43
Identities = 89/200 (44%), Positives = 119/200 (59%), Gaps = 4/200 (2%)
Frame = +3
Query: 1 MERSLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAG 60
ME ++ F+ + +++ + FY TCP IVR + S + +D +AA
Sbjct: 147 MECISYKHPFVFMLWLVLLSPLVSSQLYYNFYDTTCPNLTRIVRYNLLSAMSNDTRIAAS 326
Query: 61 LLRMHFHDCFVQGCDASVLIAGAGT---ERTAPPNLG-LRGYEVIDDAKAKVEAACPGVV 116
LLR+HFHDCFV GCD SVL+ T E+ A PN +RG+EVID K+ +E ACP V
Sbjct: 327 LLRLHFHDCFVNGCDGSVLLDDTSTQKGEKNALPNKNSIRGFEVIDTIKSALEKACPSTV 506
Query: 117 SCADILALAARDSVVLSGGLSWQVPTGRRDGRVSQASDVNNLPAPFDSVDVQKQKFAAKG 176
SCADIL LAAR++V LS G W VP GRRDG + S+ NNLP+PF+ ++ KF +KG
Sbjct: 507 SCADILTLAAREAVYLSRGPFWSVPLGRRDGTTASESEANNLPSPFEPLENITAKFISKG 686
Query: 177 LNTQDLVTLVGGHTIGTTAC 196
L +D+ L G HT G C
Sbjct: 687 LEKKDVAVLSGAHTFGFAQC 746
>TC15282 similar to UP|Q9ZNZ6 (Q9ZNZ6) Peroxidase precursor (Fragment) ,
partial (47%)
Length = 604
Score = 165 bits (417), Expect = 1e-41
Identities = 83/162 (51%), Positives = 116/162 (71%), Gaps = 3/162 (1%)
Frame = +1
Query: 3 RSLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLL 62
+S F++ L++ L +++ + H Q + GFY +TCP+AE I+ V H+ + +LAA L+
Sbjct: 100 QSCFKV--LIVCLLALIASNHAQLEQ-GFYTKTCPKAEKIILDFVHEHIHNAPSLAAALI 270
Query: 63 RMHFHDCFVQGCDASVLIAGAG--TERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCAD 120
RM+FHDCFV+GCDAS+L+ ER APPNL +RG++ ID K+ VEA CPGVVSCAD
Sbjct: 271 RMNFHDCFVRGCDASILLNSTSKQAERDAPPNLTVRGFDFIDRIKSLVEAQCPGVVSCAD 450
Query: 121 ILALAARDSVVLSGGLSWQVPTGRRDGRVSQASDV-NNLPAP 161
I+ALAARDS+V +GG W+VPTGRRDG +S + + +PAP
Sbjct: 451 IIALAARDSIVATGGPFWKVPTGRRDGVISNLVEARSQIPAP 576
>AV764870
Length = 485
Score = 161 bits (408), Expect = 1e-40
Identities = 81/108 (75%), Positives = 91/108 (84%), Gaps = 1/108 (0%)
Frame = -2
Query: 218 SFLPQLQALCPQNSGVSNRVALDTTSQNRFDTSYYANLRNGRGILLSDQALWNDASTKTF 277
SF Q+QALCPQNSG NR+ALDT SQN+FD S+YANLRNGRGIL DQALWND S KT+
Sbjct: 484 SFFLQIQALCPQNSGGWNRIALDTASQNKFDPSFYANLRNGRGILQFDQALWNDVSPKTY 305
Query: 278 VRRYLG-LRGLLGLKFNVEFGKSMVKMSNIELKTGSDSEIRKICSAFN 324
V+R LG L+GLLGL FNVEFG+SMVKM+N+ KTGSD EIRKIC AFN
Sbjct: 304 VQRDLGLLKGLLGLTFNVEFGRSMVKMNNLGFKTGSDGEIRKICFAFN 161
>TC17595 similar to UP|Q9XFI7 (Q9XFI7) Peroxidase (Fragment), partial (42%)
Length = 657
Score = 155 bits (392), Expect = 9e-39
Identities = 81/147 (55%), Positives = 104/147 (70%), Gaps = 1/147 (0%)
Frame = +1
Query: 8 IAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFH 67
+ F+L+ L S + +VGFY TCP AESIV++ V V SD +AA LLR+HFH
Sbjct: 130 VLFVLVLLFSFLIVSSDGQLQVGFYSNTCPHAESIVQAVVRGAVASDPNMAAVLLRLHFH 309
Query: 68 DCFVQGCDASVLIA-GAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAA 126
DCFV+GCD S+LI G +E+ A + G+RG+EVI+ AKA+ EA+CP VVSCADI+ALAA
Sbjct: 310 DCFVEGCDGSILIENGDQSEKLAFGHQGVRGFEVIERAKAQSEASCPDVVSCADIVALAA 489
Query: 127 RDSVVLSGGLSWQVPTGRRDGRVSQAS 153
DS+V+ G +QVPTGRRDG VS S
Sbjct: 490 TDSIVMGNGPEYQVPTGRRDGSVSNIS 570
>TC9166 similar to UP|Q9XFL4 (Q9XFL4) Peroxidase 3, partial (49%)
Length = 559
Score = 139 bits (350), Expect = 6e-34
Identities = 74/168 (44%), Positives = 103/168 (61%), Gaps = 6/168 (3%)
Frame = +3
Query: 4 SLFRIAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLR 63
S I L L + ++ + + FY + CP + V+S V S V + + LLR
Sbjct: 42 SAANIFVLSLFMLFLIGSSNSAQLSENFYVKKCPSVFNAVKSVVHSAVAKEARMGGSLLR 221
Query: 64 MHFHDCFVQGCDASVLIAGAGT---ERTAPPNLG-LRGYEVIDDAKAKVEAACPGVVSCA 119
+ FHDCFV GCD SVL+ + E+TAPPN LRG++VID K+KVEA CPGVVSCA
Sbjct: 222 LFFHDCFVNGCDGSVLLDDTSSFKGEKTAPPNSNSLRGFDVIDAIKSKVEAVCPGVVSCA 401
Query: 120 DILALAARDSVVLSGGLSWQVPTGRRDGRVSQASDVNN--LPAPFDSV 165
D++A+AARDSV + GG W+V GRRD + + + N+ +P+PF S+
Sbjct: 402 DVVAIAARDSVAILGGPYWKVKLGRRDSKTASFNAANSGVIPSPFSSL 545
>BP051104
Length = 566
Score = 136 bits (342), Expect(2) = 4e-33
Identities = 78/165 (47%), Positives = 96/165 (57%), Gaps = 5/165 (3%)
Frame = +2
Query: 71 VQGCDASVLIAGA----GTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAA 126
V GCDAS+L G + A N RG+ VID KA +E CPGVVSCAD+LALAA
Sbjct: 71 VNGCDASILXDDTSNFIGEQTAAANNRSARGFNVIDGIKANLEKQCPGVVSCADVLALAA 250
Query: 127 RDSVVLSGGLSWQVPTGRRDGRVSQASDVNN-LPAPFDSVDVQKQKFAAKGLNTQDLVTL 185
RDSVV GG SW+V GRRD + NN +P PF S+ FA +GL+ DLV L
Sbjct: 251 RDSVVQLGGPSWEVGLGRRDSTTASRGTANNTIPGPFLSLSGLITNFANQGLSVTDLVAL 430
Query: 186 VGGHTIGTTACQFFSNRLYNFTSNGPDSSIDASFLPQLQALCPQN 230
G HTIG C+ F +YN DS+IDAS+ L+ CP++
Sbjct: 431 SGAHTIGLAQCKNFRAHIYN------DSNIDASYAKFLKX*CPRS 547
Score = 21.6 bits (44), Expect(2) = 4e-33
Identities = 7/9 (77%), Positives = 7/9 (77%)
Frame = +3
Query: 61 LLRMHFHDC 69
L MHFHDC
Sbjct: 42 LTSMHFHDC 68
>TC13090 similar to UP|PER2_ARAHY (P22196) Cationic peroxidase 2 precursor
(PNPC2) , partial (23%)
Length = 484
Score = 136 bits (342), Expect = 5e-33
Identities = 69/77 (89%), Positives = 72/77 (92%), Gaps = 1/77 (1%)
Frame = +2
Query: 249 TSYYANLRNGRGILLSDQALWNDASTKTFVRRYLGL-RGLLGLKFNVEFGKSMVKMSNIE 307
TSYYANLRNGRGIL SDQALWNDASTKT+V+RYLGL RGLLGL FNVEFG+SMVKMSNI
Sbjct: 2 TSYYANLRNGRGILQSDQALWNDASTKTYVQRYLGLLRGLLGLTFNVEFGRSMVKMSNIG 181
Query: 308 LKTGSDSEIRKICSAFN 324
LKTGSD EIRKICSAFN
Sbjct: 182 LKTGSDGEIRKICSAFN 232
>TC16453 similar to UP|Q40372 (Q40372) Peroxidase precursor, partial (49%)
Length = 581
Score = 135 bits (341), Expect = 7e-33
Identities = 79/177 (44%), Positives = 111/177 (62%), Gaps = 8/177 (4%)
Frame = +2
Query: 8 IAFLLLALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFH 67
+ +L+ LA+ + G G+Y + CP+A I++S V+ + ++ + A LLR+HFH
Sbjct: 50 LVLVLVTLATFLIPSTLAGLTPGYYDKVCPQALPIIKSVVQKAINREQRIGASLLRLHFH 229
Query: 68 DCFVQGCDASVLIAGAGT---ERTAPPNL-GLRGYEVIDDAKAKVEAACP-GVVSCADIL 122
DCFV GCD S+L+ + E+TA PNL +RG+EV+D+ KA V+ AC VVSCADIL
Sbjct: 230 DCFVNGCDGSLLLDDTSSFVGEKTALPNLNSVRGFEVVDEIKAAVDRACKRPVVSCADIL 409
Query: 123 ALAARDSVVLSGGLS--WQVPTGRRDGR-VSQASDVNNLPAPFDSVDVQKQKFAAKG 176
A+AARDSV + GG +QV GRRD R SQA+ +NLP PF + F A+G
Sbjct: 410 AIAARDSVAILGGKQYWYQVRLGRRDARSASQAAANSNLPPPFLNFTQLLPAFQAQG 580
>AV414604
Length = 427
Score = 129 bits (323), Expect = 9e-31
Identities = 65/121 (53%), Positives = 83/121 (67%), Gaps = 3/121 (2%)
Frame = +2
Query: 13 LALASIVNTVHGQGSRVGFYRRTCPRAESIVRSAVESHVKSDRTLAAGLLRMHFHDCFVQ 72
LA+ + G R FYR++C +AE IV++ ++ HV S L A LLRMHFHDCFV+
Sbjct: 44 LAMFCFLGVCQGGSLRKQFYRKSCSQAEQIVKTTIQQHVSSRPELPAKLLRMHFHDCFVR 223
Query: 73 GCDASVLI---AGAGTERTAPPNLGLRGYEVIDDAKAKVEAACPGVVSCADILALAARDS 129
GCD SVL+ AG E+ A PNL L G++VID+ K +EA CP +VSCADILALAARD+
Sbjct: 224 GCDGSVLLNSTAGNTAEKDAIPNLSLSGFDVIDEIKEALEAKCPKIVSCADILALAARDA 403
Query: 130 V 130
V
Sbjct: 404 V 406
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,003,012
Number of Sequences: 28460
Number of extensions: 59693
Number of successful extensions: 389
Number of sequences better than 10.0: 99
Number of HSP's better than 10.0 without gapping: 314
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 315
length of query: 324
length of database: 4,897,600
effective HSP length: 90
effective length of query: 234
effective length of database: 2,336,200
effective search space: 546670800
effective search space used: 546670800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0314.22


