

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.7
(1046 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
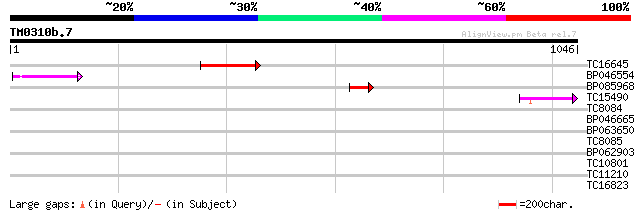
Score E
Sequences producing significant alignments: (bits) Value
TC16645 124 6e-29
BP046554 84 1e-16
BP085968 62 5e-10
TC15490 weakly similar to UP|Q9LTU4 (Q9LTU4) Helicase-like prote... 46 3e-05
TC8084 similar to UP|PIF1_YEAST (P07271) DNA repair and recombin... 40 0.002
BP046665 39 0.003
BP063650 37 0.007
TC8085 similar to GB|BAB03033.1|11994717|AP001300 AtDMC1 (meioti... 31 0.88
BP062903 30 2.0
TC10801 similar to GB|AAP42747.1|30984568|BT008734 At1g17650 {Ar... 28 9.8
TC11210 similar to GB|AAP47478.1|32331901|AY164904 RTNLB7 {Oryza... 28 9.8
TC16823 similar to UP|Q8LGA2 (Q8LGA2) Seed maturation-like prote... 28 9.8
>TC16645
Length = 596
Score = 124 bits (312), Expect = 6e-29
Identities = 57/110 (51%), Positives = 82/110 (73%)
Frame = +1
Query: 353 GVDLTYSEFPSKFVYNEKFIIWELRKQGYSIRRLTYIPPGTGELYYMRMLLAVQRGCTSF 412
G +LTY+EFPS+F+Y++ W +++ +S+ RL +I PG GE YYMR+LL +Q+GC SF
Sbjct: 34 GRNLTYAEFPSEFLYHQHTTKWVPKQKRFSLGRLPFIAPGMGENYYMRVLLTMQKGCDSF 213
Query: 413 ESIRTVKGEIHSTYQKACYALGLLSDDKQYIGAIREASELGSGNQLRGLF 462
+SI+TVKG ++ T+ AC A+GLL DD++Y+ I +SE GSG QLR LF
Sbjct: 214 KSIKTVKGVVYPTFHDACEAMGLLEDDREYVDGISISSEFGSGTQLRKLF 363
Score = 35.8 bits (81), Expect = 0.036
Identities = 16/38 (42%), Positives = 22/38 (57%)
Frame = +2
Query: 451 ELGSGNQLRGLFVSLIITNSMSRPDHVWESTWQLLAEG 488
EL GN F +++TN +SRP VW +W LL +G
Sbjct: 341 ELN*GN----FFTRMLMTNLISRPQEVWRKSWSLLCDG 442
>BP046554
Length = 556
Score = 84.0 bits (206), Expect = 1e-16
Identities = 55/129 (42%), Positives = 71/129 (54%), Gaps = 1/129 (0%)
Frame = -3
Query: 6 YKSTYHSCCLNFQEKTTAVVIKETNIPMFGFS-FIPYSQLINLSIDMFNNCQDAMAICKK 64
Y +Y+ C E+ A+ ETN G F P+ L++++I + I KK
Sbjct: 431 YWKSYYICSTQ*WEE--AIDKGETNPSSVGSELFCPHLLLVDIAICSIISRMQWPCIFKK 258
Query: 65 MGFPYLFITITCNSNWKEIADFVCSINLKPDERPYIVCRVFKMKVDNLIADLKKENIFGE 124
G+P LFIT TCNS W EI FV NL ++ P I RVFKMK+D LI+DLKK IFG
Sbjct: 257 FGYPDLFITFTCNSAWCEIQRFVQPRNLNVEDCPNICVRVFKMKLDRLISDLKKGKIFGA 78
Query: 125 VDAGLPHAH 133
DA + H H
Sbjct: 77 SDA-VRHIH 54
>BP085968
Length = 341
Score = 62.0 bits (149), Expect = 5e-10
Identities = 28/43 (65%), Positives = 36/43 (83%)
Frame = +3
Query: 628 RTAHSRFSIPLNINETLTCKVSKGSLKAKLLQQSSLIIWDEAP 670
RTAHSRFSI ++IN+ TC + +GS KA+LLQ++S IIWDEAP
Sbjct: 213 RTAHSRFSIHISINDISTCNIKQGSQKAELLQKASSIIWDEAP 341
>TC15490 weakly similar to UP|Q9LTU4 (Q9LTU4) Helicase-like protein, partial
(8%)
Length = 634
Score = 46.2 bits (108), Expect = 3e-05
Identities = 36/112 (32%), Positives = 56/112 (49%), Gaps = 6/112 (5%)
Frame = +1
Query: 941 KNLHW*NEFDSPRLWSSIQ------I*KTTVSIGCLLRNDHQQESRAVFEPCWIIPS*TS 994
+N HW F S SI +*KT+VS +L +D++ +SR+V P W I
Sbjct: 157 RNPHWR*HFHSQNGHGSI*LKLPV*V*KTSVSDQFVLCHDYK*KSRSVIVPRWPISFSPC 336
Query: 995 IHSWSVLRSSIKSQK*KSVKNPNTG**RKNYHFSNKCCLQGGL*KYLRFVLY 1046
+H+W LR S + + + + G* RKN + L +*KY+++V Y
Sbjct: 337 LHTWLALRCSPQGKIKEMT*IISAG*GRKNDQHYQERSLPRNI*KYMKYVFY 492
>TC8084 similar to UP|PIF1_YEAST (P07271) DNA repair and recombination
protein PIF1, mitochondrial precursor, partial (4%)
Length = 560
Score = 40.0 bits (92), Expect = 0.002
Identities = 35/111 (31%), Positives = 54/111 (48%), Gaps = 6/111 (5%)
Frame = +3
Query: 942 NLHW*NEFDSPRLWSSIQ------I*KTTVSIGCLLRNDHQQESRAVFEPCWIIPS*TSI 995
N HW F S SI +*K VS +L ND++ +SR+V P I I
Sbjct: 132 NQHWR*HFHSKIGHGSI*LGLPV*V*KALVSDQFVLCNDYK*KSRSVTIPR*PISFSPCI 311
Query: 996 HSWSVLRSSIKSQK*KSVKNPNTG**RKNYHFSNKCCLQGGL*KYLRFVLY 1046
H+WS + S++ + K + + G* RK+ + L +*KY+++V Y
Sbjct: 312 HTWSAVCCSLQGKIKKRTQTISAG*GRKSDKHYKERSLPRSI*KYMKYVFY 464
>BP046665
Length = 524
Score = 39.3 bits (90), Expect = 0.003
Identities = 27/69 (39%), Positives = 37/69 (53%), Gaps = 6/69 (8%)
Frame = -3
Query: 942 NLHW*NEFDSPRLWSS------IQI*KTTVSIGCLLRNDHQQESRAVFEPCWIIPS*TSI 995
N HW F S S + +*KT+VS +L ND++ +SR+V P W I I
Sbjct: 360 NQHWR*HFHSKNGHGSL*LRIPV*V*KTSVSDQFVLCNDYK*KSRSVTIPRWPISFSPCI 181
Query: 996 HSWSVLRSS 1004
H+WSV+R S
Sbjct: 180 HTWSVVRCS 154
>BP063650
Length = 497
Score = 37.0 bits (84), Expect(2) = 0.007
Identities = 17/32 (53%), Positives = 24/32 (74%)
Frame = +1
Query: 713 QILHVIQKGSRQDIVTATVNSSYLWENCKVLS 744
+ L V+ KG QDI+ A VN+SYL ++C+VLS
Sbjct: 58 KFLTVVYKGRMQDIIHAIVNASYL*DHCQVLS 153
Score = 20.0 bits (40), Expect(2) = 0.007
Identities = 8/12 (66%), Positives = 10/12 (82%)
Frame = +3
Query: 705 VVLGGDFKQILH 716
VVL GDF+QI +
Sbjct: 33 VVLQGDFRQIFN 68
>TC8085 similar to GB|BAB03033.1|11994717|AP001300 AtDMC1 (meiotic
recombination protein)-like protein {Arabidopsis
thaliana;} , partial (8%)
Length = 500
Score = 31.2 bits (69), Expect = 0.88
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = +1
Query: 471 MSRPDHVWESTWQLLA 486
M+ PDHVW TW+LLA
Sbjct: 115 MTNPDHVWNITWKLLA 162
>BP062903
Length = 554
Score = 30.0 bits (66), Expect = 2.0
Identities = 14/36 (38%), Positives = 19/36 (51%), Gaps = 6/36 (16%)
Frame = +3
Query: 60 AICKKMGFPYLF------ITITCNSNWKEIADFVCS 89
A C + G PY+F +C WKEI+ +VCS
Sbjct: 198 ARCPRQGSPYVF*XQFRTSRTSCELRWKEISIYVCS 305
>TC10801 similar to GB|AAP42747.1|30984568|BT008734 At1g17650 {Arabidopsis
thaliana;}, partial (26%)
Length = 565
Score = 27.7 bits (60), Expect = 9.8
Identities = 17/63 (26%), Positives = 30/63 (46%)
Frame = -2
Query: 26 IKETNIPMFGFSFIPYSQLINLSIDMFNNCQDAMAICKKMGFPYLFITITCNSNWKEIAD 85
I + N+P+ S + ++ L + FN+ + + I K MGF I+C NW + +
Sbjct: 330 IFQVNLPLIRRSLV--F*VLKLGLQCFNDS*EVL-ITKAMGFCNFI*LISCCCNWNRLGN 160
Query: 86 FVC 88
C
Sbjct: 159 RFC 151
>TC11210 similar to GB|AAP47478.1|32331901|AY164904 RTNLB7 {Oryza sativa
(japonica cultivar-group);} , partial (7%)
Length = 602
Score = 27.7 bits (60), Expect = 9.8
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 2/36 (5%)
Frame = -2
Query: 158 LPDPQLYPKLYDVVST*IMHGPCGKANPT--CACMK 191
L PQ P D + IMHGPC K PT +C+K
Sbjct: 373 LGKPQQTPHRQDSIGINIMHGPC-KQTPTSFTSCLK 269
>TC16823 similar to UP|Q8LGA2 (Q8LGA2) Seed maturation-like protein, partial
(29%)
Length = 711
Score = 27.7 bits (60), Expect = 9.8
Identities = 23/73 (31%), Positives = 38/73 (51%)
Frame = +3
Query: 726 IVTATVNSSYLWENCKVLSLTKNMRLLNTNGHQDNDDVKEFSE*ILKLGNGESTPNDDGK 785
+V++ + LW +SLT+N+ + + G +D+D K SE + G GE D G+
Sbjct: 432 LVSSIITGYTLWNAEYRMSLTRNLDIASPCGARDSDCEKR-SEILEVKGGGE----DGGE 596
Query: 786 MLIDVPHDLFITD 798
I+V DL + D
Sbjct: 597 --IEVASDLGLKD 629
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.333 0.145 0.469
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,893,804
Number of Sequences: 28460
Number of extensions: 295722
Number of successful extensions: 2136
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 2107
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2135
length of query: 1046
length of database: 4,897,600
effective HSP length: 100
effective length of query: 946
effective length of database: 2,051,600
effective search space: 1940813600
effective search space used: 1940813600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0310b.7


