

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0310b.14
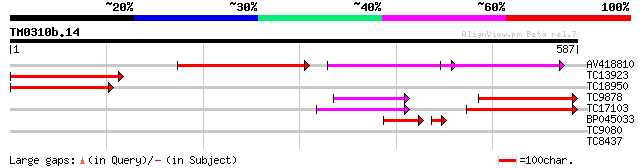
(587 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV418810 257 3e-69
TC13923 homologue to UP|Q38950 (Q38950) Protein phosphatase 2A 6... 220 6e-58
TC18950 homologue to UP|Q9FVD6 (Q9FVD6) Ser/Thr specific protein... 211 3e-55
TC9878 homologue to UP|2AAA_PEA (P36875) Protein phosphatase PP2... 195 2e-50
TC17103 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protein... 184 3e-47
BP045033 52 1e-10
TC9080 28 5.4
TC8437 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate dehy... 27 9.3
>AV418810
Length = 415
Score = 257 bits (657), Expect = 3e-69
Identities = 127/137 (92%), Positives = 134/137 (97%)
Frame = +3
Query: 174 DDMPMVRRSAATNLGKFAATVEAAHLKSDIMSVFDDLTQDDQDSVRLLAVEGCAALGKLL 233
DDMPMVRRSAA+NLGKFAATVE AHLK+DIMS+F+DLT+DDQDSVRLLAVEGCAALGKLL
Sbjct: 3 DDMPMVRRSAASNLGKFAATVEYAHLKADIMSIFEDLTKDDQDSVRLLAVEGCAALGKLL 182
Query: 234 EPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNE 293
EPQ+C+ HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNE
Sbjct: 183 EPQDCITHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNE 362
Query: 294 AEVRIAAAGKVTKFSRI 310
AEVRIAAAGKVTKF RI
Sbjct: 363 AEVRIAAAGKVTKFCRI 413
Score = 44.3 bits (103), Expect = 6e-05
Identities = 31/128 (24%), Positives = 57/128 (44%)
Frame = +3
Query: 447 DKVYSIRDAAANNVKRLAEEFGPDWAMQHIIPQVLDMINDPHYLYRMTILQAISLLAPVL 506
D + +R +AA+N+ + A I+ D+ D R+ ++ + L +L
Sbjct: 3 DDMPMVRRSAASNLGKFAATVEYAHLKADIMSIFEDLTKDDQDSVRLLAVEGCAALGKLL 182
Query: 507 GSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVENTLRPCLVELSEDPD 566
+ + +LP++VN S+D+ +++ VA L L V L P V L D +
Sbjct: 183 EPQDCITHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNE 362
Query: 567 VDVRFFAS 574
+VR A+
Sbjct: 363 AEVRIAAA 386
Score = 42.4 bits (98), Expect = 2e-04
Identities = 33/133 (24%), Positives = 57/133 (42%)
Frame = +3
Query: 330 DSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVI 389
D VR + AS + A + ++ IF L KD+ VRL + + +++
Sbjct: 3 DDMPMVRRSAASNLGKFAATVEYAHLKADIMSIFEDLTKDDQDSVRLLAVEGCAALGKLL 182
Query: 390 GIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKV 449
+LP IV ++D+ WRVR + + L +G +L ++ L+D
Sbjct: 183 EPQDCITHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNE 362
Query: 450 YSIRDAAANNVKR 462
+R AAA V +
Sbjct: 363 AEVRIAAAGKVTK 401
>TC13923 homologue to UP|Q38950 (Q38950) Protein phosphatase 2A 65 kDa
regulatory subunit, partial (20%)
Length = 515
Score = 220 bits (560), Expect = 6e-58
Identities = 109/118 (92%), Positives = 116/118 (97%)
Frame = +1
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
M+MVD+PLYPIAVLIDELKN+DIQLRLNSIR+LSTIARALGEERTRKELIPFL ENNDDD
Sbjct: 160 MSMVDEPLYPIAVLIDELKNDDIQLRLNSIRKLSTIARALGEERTRKELIPFLGENNDDD 339
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSVESLCRIGAQMR 118
DEVLLAMAEELGVFIP+VGGVEHA+VLLPPLETLCTVEETCVRDK+VESLCRIG QMR
Sbjct: 340 DEVLLAMAEELGVFIPFVGGVEHAHVLLPPLETLCTVEETCVRDKAVESLCRIGPQMR 513
Score = 28.1 bits (61), Expect = 4.2
Identities = 22/76 (28%), Positives = 38/76 (49%), Gaps = 2/76 (2%)
Frame = +1
Query: 357 EQLLPIFLSL--LKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
E L PI + + LK++ +RLN I KL + + +G + + L+P + E +D
Sbjct: 175 EPLYPIAVLIDELKNDDIQLRLNSIRKLSTIARALGEERTRKELIPFLGENNDDDD---- 342
Query: 415 LAIIEYIPLLASQLGV 430
E + +A +LGV
Sbjct: 343 ----EVLLAMAEELGV 378
>TC18950 homologue to UP|Q9FVD6 (Q9FVD6) Ser/Thr specific protein
phosphatase 2A A regulatory subunit beta isoform,
partial (18%)
Length = 599
Score = 211 bits (537), Expect = 3e-55
Identities = 107/107 (100%), Positives = 107/107 (100%)
Frame = +3
Query: 1 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 60
MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD
Sbjct: 279 MAMVDQPLYPIAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDD 458
Query: 61 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSV 107
DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSV
Sbjct: 459 DEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETCVRDKSV 599
Score = 28.5 bits (62), Expect = 3.2
Identities = 18/70 (25%), Positives = 36/70 (50%)
Frame = +3
Query: 360 LPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVRLAIIE 419
+ + + LK+E +RLN I +L + + +G + + L+P + E +D V LA+ E
Sbjct: 309 IAVLIDELKNEDIQLRLNSIRRLSTIARALGEERTRKELIPFLSENNDDDD-EVLLAMAE 485
Query: 420 YIPLLASQLG 429
+ + +G
Sbjct: 486 ELGVFIPYVG 515
>TC9878 homologue to UP|2AAA_PEA (P36875) Protein phosphatase PP2A
regulatory subunit A (PR65) (Fragment), partial (26%)
Length = 617
Score = 195 bits (495), Expect = 2e-50
Identities = 101/102 (99%), Positives = 102/102 (99%)
Frame = +1
Query: 486 DPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIV 545
DPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIV
Sbjct: 1 DPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIV 180
Query: 546 EESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
EESVVENT+RPCLVELSEDPDVDVRFFASQALQSSDQVKMSS
Sbjct: 181 EESVVENTIRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 306
Score = 50.8 bits (120), Expect = 6e-07
Identities = 24/79 (30%), Positives = 44/79 (55%)
Frame = +1
Query: 336 RSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVIGIDLLS 395
R + I +APVLG + T +LLP+ ++ KD P+++ N+ L + ++ ++
Sbjct: 19 RMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVE 198
Query: 396 QSLLPAIVELAEDRHWRVR 414
++ P +VEL+ED VR
Sbjct: 199 NTIRPCLVELSEDPDVDVR 255
Score = 37.4 bits (85), Expect = 0.007
Identities = 21/83 (25%), Positives = 40/83 (47%)
Frame = +1
Query: 219 RLLAVEGCAALGKLLEPQECVAHILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTR 278
R+ ++ + L +L + + +LP++VN S+D+ +++ VA L L V
Sbjct: 19 RMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIVEESVVE 198
Query: 279 TELVPAYVRLLRDNEAEVRIAAA 301
+ P V L D + +VR A+
Sbjct: 199 NTIRPCLVELSEDPDVDVRFFAS 267
Score = 33.9 bits (76), Expect = 0.076
Identities = 21/85 (24%), Positives = 35/85 (40%)
Frame = +1
Query: 252 DKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAAAGKVTKFSRIL 311
D + R + + L +G E T ++L+P V +D ++ A + I+
Sbjct: 1 DPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIV 180
Query: 312 SPELAIEHILPCVKELSADSSQHVR 336
+ I PC+ ELS D VR
Sbjct: 181 EESVVENTIRPCLVELSEDPDVDVR 255
Score = 33.5 bits (75), Expect = 0.099
Identities = 26/92 (28%), Positives = 41/92 (44%)
Frame = +1
Query: 408 DRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDAAANNVKRLAEEF 467
D H+ R+ I++ I LLA LG KL L + KD+V +I+ A ++ L
Sbjct: 1 DPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFNVAKVLQSLIPIV 180
Query: 468 GPDWAMQHIIPQVLDMINDPHYLYRMTILQAI 499
I P ++++ DP R QA+
Sbjct: 181 EESVVENTIRPCLVELSEDPDVDVRFFASQAL 276
>TC17103 homologue to UP|Q9FVD7 (Q9FVD7) Ser/Thr specific protein
phosphatase 2A A regulatory subunit alpha isoform,
partial (19%)
Length = 685
Score = 184 bits (467), Expect = 3e-47
Identities = 91/114 (79%), Positives = 107/114 (93%)
Frame = +2
Query: 474 QHIIPQVLDMINDPHYLYRMTILQAISLLAPVLGSEITSSKLLPLVVNASKDRVPNIKFN 533
Q+IIPQVLDMIN+PHYLYRMTIL+AISLLAPV+GSEIT SKLLP+VV+ASKDRVPNIKFN
Sbjct: 2 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 181
Query: 534 VAKVLQSLIPIVEESVVENTLRPCLVELSEDPDVDVRFFASQALQSSDQVKMSS 587
VAKVL+S+ PI+++SVVE T+RPCLVELSEDPDVDVRFF++ ALQ+ D V MS+
Sbjct: 182 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVMMST 343
Score = 57.0 bits (136), Expect = 8e-09
Identities = 27/97 (27%), Positives = 55/97 (55%)
Frame = +2
Query: 318 EHILPCVKELSADSSQHVRSALASVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLN 377
++I+P V ++ + R + I +APV+G + T +LLP+ +S KD P+++ N
Sbjct: 2 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 181
Query: 378 IISKLDQVNQVIGIDLLSQSLLPAIVELAEDRHWRVR 414
+ L+ + ++ ++ +++ P +VEL+ED VR
Sbjct: 182 VAKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 292
Score = 38.5 bits (88), Expect = 0.003
Identities = 23/96 (23%), Positives = 43/96 (43%)
Frame = +2
Query: 241 HILPVIVNFSQDKSWRVRYMVANQLYELCEAVGPEPTRTELVPAYVRLLRDNEAEVRIAA 300
+I+P +++ + + R + + L +G E T ++L+P V +D ++
Sbjct: 5 NIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNV 184
Query: 301 AGKVTKFSRILSPELAIEHILPCVKELSADSSQHVR 336
A + IL + + I PC+ ELS D VR
Sbjct: 185 AKVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVR 292
Score = 36.6 bits (83), Expect = 0.012
Identities = 20/109 (18%), Positives = 49/109 (44%)
Frame = +2
Query: 281 LVPAYVRLLRDNEAEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALA 340
++P + ++ + R+ ++ + ++ E+ +LP V S D +++ +A
Sbjct: 8 IIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFNVA 187
Query: 341 SVIMGMAPVLGKDATIEQLLPIFLSLLKDEFPDVRLNIISKLDQVNQVI 389
V+ + P+L + + + P + L +D DVR L ++ V+
Sbjct: 188 KVLESIFPILDQSVVEKTIRPCLVELSEDPDVDVRFFSNPALQAIDHVM 334
Score = 35.0 bits (79), Expect = 0.034
Identities = 21/92 (22%), Positives = 46/92 (49%)
Frame = +2
Query: 396 QSLLPAIVELAEDRHWRVRLAIIEYIPLLASQLGVGFFDDKLGALCMQWLKDKVYSIRDA 455
Q+++P ++++ + H+ R+ I+ I LLA +G KL + + KD+V +I+
Sbjct: 2 QNIIPQVLDMINNPHYLYRMTILRAISLLAPVMGSEITCSKLLPVVVSASKDRVPNIKFN 181
Query: 456 AANNVKRLAEEFGPDWAMQHIIPQVLDMINDP 487
A ++ + + I P ++++ DP
Sbjct: 182 VAKVLESIFPILDQSVVEKTIRPCLVELSEDP 277
>BP045033
Length = 434
Score = 52.4 bits (124), Expect(2) = 1e-10
Identities = 28/41 (68%), Positives = 32/41 (77%)
Frame = -3
Query: 388 VIGIDLLSQSLLPAIVELAEDRHWRVRLAIIEYIPLLASQL 428
VIGI+ LSQSLLP + ELAEDRH RV LAI +IPLLA +
Sbjct: 300 VIGIN*LSQSLLPPLAELAEDRHLRVWLAITVHIPLLAKPI 178
Score = 30.4 bits (67), Expect(2) = 1e-10
Identities = 11/16 (68%), Positives = 13/16 (80%)
Frame = -1
Query: 437 LGALCMQWLKDKVYSI 452
LGALCMQWL D+ Y +
Sbjct: 176 LGALCMQWLLDQAYEV 129
>TC9080
Length = 620
Score = 27.7 bits (60), Expect = 5.4
Identities = 19/89 (21%), Positives = 37/89 (41%)
Frame = +1
Query: 294 AEVRIAAAGKVTKFSRILSPELAIEHILPCVKELSADSSQHVRSALASVIMGMAPVLGKD 353
A+ ++ + V+K E + ++ +L D R A SV + L KD
Sbjct: 64 AKAAVSLSNCVSKMGVEEMEEFGMGEMIEVAADLVNDRLPEARDAARSVATTVYEALTKD 243
Query: 354 ATIEQLLPIFLSLLKDEFPDVRLNIISKL 382
A +EQ + ++ S + + + I K+
Sbjct: 244 AEVEQKMEVWQSFCQSKLQPIHALSILKI 330
>TC8437 homologue to GB|AAB86804.1|2454184|ATU80186 pyruvate dehydrogenase
E1 beta subunit {Arabidopsis thaliana;} , partial (40%)
Length = 915
Score = 26.9 bits (58), Expect = 9.3
Identities = 24/102 (23%), Positives = 43/102 (41%), Gaps = 3/102 (2%)
Frame = +1
Query: 3 MVDQPLYPIAVLIDELKNEDIQLRLNSIR---RLSTIARALGEERTRKELIPFLSENNDD 59
+V++ P + I LK D+ NS++ R+ + + L +SEN +D
Sbjct: 184 LVNKGYDPEVIDIRSLKPFDLHTIGNSVKKTHRVLIVEECMRTGGIGASLTAAISENFND 363
Query: 60 DDEVLLAMAEELGVFIPYVGGVEHANVLLPPLETLCTVEETC 101
+ + V PY G +E V + P + + VE+ C
Sbjct: 364 YLDAPIVCLSSQDVPTPYAGPLEEMTV-VQPAQIVTAVEQLC 486
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,818,305
Number of Sequences: 28460
Number of extensions: 110555
Number of successful extensions: 475
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 462
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 474
length of query: 587
length of database: 4,897,600
effective HSP length: 95
effective length of query: 492
effective length of database: 2,193,900
effective search space: 1079398800
effective search space used: 1079398800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0310b.14


