

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0304.11
(1797 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
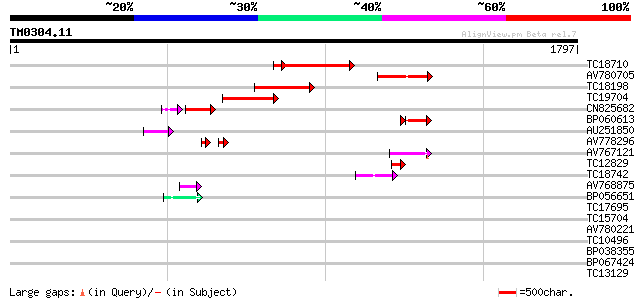
Score E
Sequences producing significant alignments: (bits) Value
TC18710 242 4e-64
AV780705 191 1e-48
TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, p... 188 6e-48
TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA po... 164 1e-40
CN825682 120 1e-30
BP060613 96 1e-20
AU251850 72 8e-13
AV778296 45 7e-11
AV767121 55 7e-08
TC12829 47 3e-05
TC18742 similar to UP|Q9FZG5 (Q9FZG5) T2E6.4, partial (3%) 45 8e-05
AV768875 44 2e-04
BP056651 42 9e-04
TC17695 homologue to UP|Q41891 (Q41891) ZEM1 (Fragment), partial... 37 0.021
TC15704 similar to UP|O22265 (O22265) Expressed protein (At2g474... 36 0.061
AV780221 35 0.080
TC10496 35 0.14
BP038355 27 0.19
BP067424 31 1.5
TC13129 similar to UP|RIR2_TOBAC (P49730) Ribonucleoside-diphosp... 31 2.0
>TC18710
Length = 843
Score = 242 bits (617), Expect = 4e-64
Identities = 117/234 (50%), Positives = 165/234 (70%)
Frame = +2
Query: 860 EEIKVVAHSMDSLKAPGPDGLNGLFFKTHWNVVGPEVCSVVHSFFNTSTLSPCINETLVA 919
++++++ +++ + G DG+NGLF++ + +VV V + +F + + INETLV
Sbjct: 140 KKLRLLFFLLETSQPRGMDGMNGLFYQKNGDVVKDSVT*AILAFLERAEIPNEINETLVT 319
Query: 920 LVPKIPSLEMANQYRPISCCNYLYKIISKVIVARLKGELNDIITPNQSAFIKDRLIQDNI 979
L+PK+P E NQ+RPIS C +LYK+ISK+ VARLK + D+I+P QS FI+ R IQDN+
Sbjct: 320 LIPKVPHPESINQFRPISGCTFLYKVISKIFVARLKDAMPDLISPMQSGFIQGRQIQDNL 499
Query: 980 LIAHEIFHALQHRGREHKDHLAIKLDMSKAYDRVEWEFLEKALLAYGFCGPWVSLVMNLV 1039
LI E FHA+ G ++H IKLDM+KAYDRVEW+FLE +LLA+GF WV ++M LV
Sbjct: 500 LIVQEAFHAINRPGALGRNHSIIKLDMNKAYDRVEWKFLESSLLAFGFSTNWVKMIMILV 679
Query: 1040 RGVSYKYKVNGYLSRKYIPQRGLRQGDPLSPYLFVLVMDVLSHMILQAKNQGHI 1093
GVSY YK+NG + K +PQRGLRQGDP SPYLF+ M+VLS +I + N G++
Sbjct: 680 SGVSYNYKINGVVGPKLLPQRGLRQGDPFSPYLFLFTMEVLSLLIQNSFNMGNL 841
Score = 47.8 bits (112), Expect = 2e-05
Identities = 23/42 (54%), Positives = 26/42 (61%)
Frame = +1
Query: 835 QECLSCVPKLIPTDLNNKLMETVTDEEIKVVAHSMDSLKAPG 876
+ECL VP L+ DLN KL VT EEIK V S+ L APG
Sbjct: 64 RECLDVVPSLVSQDLNQKLESQVTSEEIKAVVFSLGDLTAPG 189
>AV780705
Length = 524
Score = 191 bits (484), Expect = 1e-48
Identities = 89/178 (50%), Positives = 120/178 (67%), Gaps = 5/178 (2%)
Frame = -2
Query: 1166 NIFQIPTWEEPGHYLGLPAVWGRSKTRALAWIKERMKEKIAGWKEVLLNQAGKEVLIKSV 1225
N+ +P W++PGHYLGLPA+WGR+K+ +L WI+E++KEK+ GWKE LLNQAGKEVLIK++
Sbjct: 523 NLLHMPIWDDPGHYLGLPAIWGRNKSHSLVWIEEKVKEKLEGWKETLLNQAGKEVLIKAI 344
Query: 1226 IQSIPTYAMAVTPFPKGFCKELDSLVARFWWSNNKKSRGIHLKDWHSLT-----CSKKEG 1280
IQ+IP+YAM + FPK FC L +LVA FWW KS+G ++ SK++G
Sbjct: 343 IQAIPSYAMTIVHFPKTFCNSLYALVADFWW----KSQGEGAVEYTGAVGTK*PLSKEKG 176
Query: 1281 GMGFREFHLQNLAHMTKQAWRIHNTPHTLVAQVMKALYFPDSSFLDATPKPGDSWMWR 1338
G+GFR+ QNLA + +QAWR+ P L +VMK+LYFP + WMWR
Sbjct: 175 GVGFRDLRTQNLAFLARQAWRVLTNPEALWVRVMKSLYFPTPALCLLLWDGTHPWMWR 2
>TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, partial
(14%)
Length = 592
Score = 188 bits (478), Expect = 6e-48
Identities = 91/188 (48%), Positives = 130/188 (68%)
Frame = -3
Query: 777 GDRNTHFFHATTIQRRGRNRIHRLKDGTGSWIEGQEGVMENITDHFQLIFKSEQVTQIQE 836
GD NT FFHA+TIQRR NRI ++KD G W+EGQ+ V E ++F I+ S + ++E
Sbjct: 569 GDHNTRFFHASTIQRRDFNRILKIKDAHGIWVEGQKKVNEAAVNYFSDIYSSSPLQYLRE 390
Query: 837 CLSCVPKLIPTDLNNKLMETVTDEEIKVVAHSMDSLKAPGPDGLNGLFFKTHWNVVGPEV 896
CL+ +PKL+ LN+KL V+ +EIK SM LKAPG DG+NGLF++ + +V +V
Sbjct: 389 CLAPIPKLVTDRLNHKLCAQVSTDEIKEAVFSMGDLKAPGMDGINGLFYQQNREIVKNDV 210
Query: 897 CSVVHSFFNTSTLSPCINETLVALVPKIPSLEMANQYRPISCCNYLYKIISKVIVARLKG 956
+ + FF+ L +NETLV L+PKIP E Q+RPISCC+++YK+ISK+IVARLK
Sbjct: 209 NNAILDFFDHGELPVELNETLVTLIPKIPHAEAIQQFRPISCCSFIYKVISKIIVARLKP 30
Query: 957 ELNDIITP 964
++ ++I+P
Sbjct: 29 DMVNLISP 6
>TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA
polymerase-like protein, partial (3%)
Length = 530
Score = 164 bits (414), Expect = 1e-40
Identities = 76/176 (43%), Positives = 111/176 (62%), Gaps = 1/176 (0%)
Frame = -3
Query: 676 DCSKVVEDAW-NTTPQGENDWETLQNKVKSCQIGLTKWQKVTFSRADKEIEKLKARLSSM 734
+C + ++ AW NT+ + L K + ++ LT+W FSRADKEI +LK +L +
Sbjct: 528 ECKETIKKAWDNTSETASSHCSNLSGKTTNTKLALTEWHHQNFSRADKEIAELKNQLEHL 349
Query: 735 HNEDHHKIDWNEMAQFKAQIRQLWRQEELFWSTRSRLQWAQWGDRNTHFFHATTIQRRGR 794
N + DW E+ Q K +++ LWRQEELFWS RSR++W + GD+NT FFHA+T+QRR R
Sbjct: 348 QNYSNSGEDWQEIKQRKMRLKDLWRQEELFWSQRSRIKWLKSGDKNTKFFHASTVQRRER 169
Query: 795 NRIHRLKDGTGSWIEGQEGVMENITDHFQLIFKSEQVTQIQECLSCVPKLIPTDLN 850
NRI R+KD G+W+ Q + ++ D + I+KS I ECL+ VP++I LN
Sbjct: 168 NRIERIKDMQGTWVRSQLAINRDVPDFYPDIYKSTNRQNITECLNAVPRVIYESLN 1
>CN825682
Length = 626
Score = 120 bits (302), Expect(2) = 1e-30
Identities = 51/95 (53%), Positives = 68/95 (70%)
Frame = -3
Query: 556 WCCLGDFNETLYQHEKDGLHPQSQQSLNRFRSFVDSNALANIDIKGCKYTWLSNPREGFV 615
W C GDFNE L+ EK+G Q + FR F+D L ++++KGC++TWLSN R G V
Sbjct: 300 WTCYGDFNEILHHQEKEGARDQDPTRIQVFRDFLDGANLMDLELKGCRFTWLSNLRNGLV 121
Query: 616 TREKLDRVLSNWSWRSLYPHALVVALPVVSSDHSP 650
T+E+LDRVL NW+WR YP+A +ALP++SSDHSP
Sbjct: 120 TKERLDRVLVNWTWRLDYPNATAMALPIISSDHSP 16
Score = 31.2 bits (69), Expect(2) = 1e-30
Identities = 24/67 (35%), Positives = 33/67 (48%)
Frame = -1
Query: 482 VDPIGTAGGLALFWNKECTVEILDSNQNYIHVMITEKEKVIGAEYTFIYGEPLRQRRQAL 541
+DPI L+W +C D Q HV KE V+G TFIY P+ ++R+ L
Sbjct: 485 IDPI------RLWWPIKCVKH--DKIQPQPHVW-PNKEIVLG---TFIYANPIFRQRRNL 342
Query: 542 WDSISRL 548
W I+ L
Sbjct: 341 WPQITSL 321
>BP060613
Length = 378
Score = 95.5 bits (236), Expect(2) = 1e-20
Identities = 42/83 (50%), Positives = 56/83 (66%)
Frame = +1
Query: 1255 WWSNNKKSRGIHLKDWHSLTCSKKEGGMGFREFHLQNLAHMTKQAWRIHNTPHTLVAQVM 1314
WWS+ K +RG+H + W LT K EGG+GF++F +QN A + KQAWRI + P L Q++
Sbjct: 46 WWSS-KGTRGVHWRKWDLLTELKDEGGLGFKDFEIQNQALLAKQAWRILHNPDALWVQIL 222
Query: 1315 KALYFPDSSFLDATPKPGDSWMW 1337
KALYFP FL T + G SW+W
Sbjct: 223 KALYFPHHDFLQTTKRTGASWVW 291
Score = 22.7 bits (47), Expect(2) = 1e-20
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = +2
Query: 1240 PKGFCKELDSLVARF 1254
PK CK+L S VARF
Sbjct: 2 PKTXCKDLCSRVARF 46
>AU251850
Length = 343
Score = 72.0 bits (175), Expect = 8e-13
Identities = 34/101 (33%), Positives = 58/101 (56%), Gaps = 4/101 (3%)
Frame = +2
Query: 423 SMRLISWNCRGVGASTTARELKEINSKQKPSILFLMETRARRNKIERLKFALHFSNFFCV 482
+M+L+SWNCRG+G+ L+ + K+ P ++FLMET+ ++ + E++ SN V
Sbjct: 35 AMKLLSWNCRGLGSPRAVEALQRLVRKEIPQLVFLMETKLKKFETEKISLGFGLSNCLIV 214
Query: 483 DPIG----TAGGLALFWNKECTVEILDSNQNYIHVMITEKE 519
D +G GGLA++WN V ++ + N+I V E +
Sbjct: 215 DCVGDEKERKGGLAMWWNASVDVSLVSFSGNHIVVACVEDD 337
>AV778296
Length = 408
Score = 45.4 bits (106), Expect(2) = 7e-11
Identities = 17/29 (58%), Positives = 25/29 (85%)
Frame = +3
Query: 608 SNPREGFVTREKLDRVLSNWSWRSLYPHA 636
S P GFVTREK++RV+++W WR+L+P+A
Sbjct: 144 SKPGGGFVTREKIERVVADWEWRALFPNA 230
Score = 40.0 bits (92), Expect(2) = 7e-11
Identities = 16/34 (47%), Positives = 22/34 (64%)
Frame = +2
Query: 661 SGRQFKYEAFWNEHKDCSKVVEDAWNTTPQGEND 694
SG QFKYE +EH +C +V+ AW TP +N+
Sbjct: 305 SGVQFKYERVGDEHVECKEVINKAWAETPDMDNE 406
Score = 35.8 bits (81), Expect = 0.061
Identities = 23/69 (33%), Positives = 36/69 (51%), Gaps = 9/69 (13%)
Frame = +1
Query: 599 IKGCKYTWLSNPREGFVTREK----LDRVLSNWSWRS----LYPHAL-VVALPVVSSDHS 649
+ CK+ S + ++ + L+R LS W W++ LY L A P +SSDHS
Sbjct: 94 LSSCKHYVCSRQKTQYLASQGGVL*LERRLSGW-WQTGNGELYSLMLKATAYPPISSDHS 270
Query: 650 PLILDPVPK 658
P++ +P PK
Sbjct: 271 PMVFEPSPK 297
>AV767121
Length = 525
Score = 55.5 bits (132), Expect = 7e-08
Identities = 35/140 (25%), Positives = 63/140 (45%), Gaps = 9/140 (6%)
Frame = +2
Query: 1205 IAGWKEVLLNQAGKEVLIKSVIQSIPTYAMAVTPFPKGFCKELDSLVARFWWSNNKKSRG 1264
++ WK+ ++ G+ LI+SV+ ++P + ++ P G K L+ F W ++
Sbjct: 2 LSKWKQKTVSFGGRIFLIQSVLTALPLFFLSFFKLPIGVGKSCVRLMRNFLWGGSENENK 181
Query: 1265 IHLKDWHSLTCSKKEGGMGFREFHLQNLAHMTKQAWRIHNTPHTLVAQVMKALYFPD--- 1321
I W + K+ GG+G ++ N A + K WR P +L +V++A PD
Sbjct: 182 IA*VKWTDVCKPKELGGLGIKDLFTFNKALLGK*RWRYLTEPDSLWRRVIEAQ--PDHYS 355
Query: 1322 ------SSFLDATPKPGDSW 1335
+ L P+ D W
Sbjct: 356 CGSSWWNDILSLCPEDADGW 415
>TC12829
Length = 448
Score = 47.0 bits (110), Expect = 3e-05
Identities = 20/46 (43%), Positives = 31/46 (66%)
Frame = +3
Query: 1210 EVLLNQAGKEVLIKSVIQSIPTYAMAVTPFPKGFCKELDSLVARFW 1255
E L +AG+EVLIKSV ++IPTY M+ P C +++ +V +F+
Sbjct: 252 EKFLYRAGREVLIKSVTKAIPTYIMSCFALPDSICSQIEGMVGKFY 389
>TC18742 similar to UP|Q9FZG5 (Q9FZG5) T2E6.4, partial (3%)
Length = 912
Score = 45.4 bits (106), Expect = 8e-05
Identities = 37/140 (26%), Positives = 64/140 (45%), Gaps = 7/140 (5%)
Frame = -3
Query: 1097 KLSPSTP---C----ITHLFFADDSLVFAKATQQEAYHFVQLLNTFSSASSQRINTSKSS 1149
++SP +P C I+HL FAD+ +VF+ + +L+ F S N +KS
Sbjct: 418 RISPQSPHWRCQNEEISHLCFADNLMVFSNGYLESIAIINNVLHIFQHLSGLTPNPAKSE 239
Query: 1150 MSTSKHIPTLLKTQMTNIFQIPTWEEPGHYLGLPAVWGRSKTRALAWIKERMKEKIAGWK 1209
+ + K Q+TN+ + P LG+P + + K + +R K+ W
Sbjct: 238 VFIL--VKETTKNQITNMLGYKEGKLPVR*LGVPQLTTKLKASDCKVLVDRKPSKLQHWT 65
Query: 1210 EVLLNQAGKEVLIKSVIQSI 1229
L+ AG+ LI S++ S+
Sbjct: 64 GRSLSYAGRLQLINSILFSM 5
>AV768875
Length = 232
Score = 44.3 bits (103), Expect = 2e-04
Identities = 23/72 (31%), Positives = 36/72 (49%), Gaps = 2/72 (2%)
Frame = +1
Query: 537 RRQALWDSISRLKPLAQGPWCCLGDFNETLYQHEKDG--LHPQSQQSLNRFRSFVDSNAL 594
+R LW +S ++ PW +GD NE L+ + G HP Q RF + +D L
Sbjct: 1 QRDILWQHLSTIRASISIPWLLIGDMNEILFPSQVRGGEFHPNRAQ---RFATVLDDCNL 171
Query: 595 ANIDIKGCKYTW 606
++ + G K+TW
Sbjct: 172 IDLGMVGGKFTW 207
>BP056651
Length = 564
Score = 42.0 bits (97), Expect = 9e-04
Identities = 30/128 (23%), Positives = 51/128 (39%), Gaps = 5/128 (3%)
Frame = +1
Query: 488 AGGLALFWNKEC-TVEILDSNQNYIHVMITEKEKVIGAEYTFIYGEPLRQRRQALWDSIS 546
AGGL W TVE + ++ + K K + +Y R ++ LW+ +
Sbjct: 106 AGGLLCVWKPSVFTVEECFLGEGFLG--LKGKWKGVSCVIINVYSPCSRGEKRQLWEDLK 279
Query: 547 RLKP-LAQGPWCCLGDFNETLYQHEKDGLHPQSQQ---SLNRFRSFVDSNALANIDIKGC 602
L+ L + WC GDFN E+ G H + + F F+D
Sbjct: 280 ALRRRLEEDIWCMAGDFNSVRRSEERRGTHGEGSSGAGEMEEFNDFID------------ 423
Query: 603 KYTWLSNP 610
++ W+++P
Sbjct: 424 EWNWMTSP 447
>TC17695 homologue to UP|Q41891 (Q41891) ZEM1 (Fragment), partial (6%)
Length = 510
Score = 37.4 bits (85), Expect = 0.021
Identities = 22/52 (42%), Positives = 25/52 (47%), Gaps = 7/52 (13%)
Frame = -1
Query: 398 IPQRTFLSKFCYH-------GWGGGH*SAPNVSMRLISWNCRGVGASTTARE 442
IPQ T + C G GGG PN M L SWNCRG+ A+ T E
Sbjct: 156 IPQETTAAIPCRRVKFYN*KG*GGGPIHGPNA*MSLFSWNCRGLAAAATIDE 1
>TC15704 similar to UP|O22265 (O22265) Expressed protein
(At2g47450/T30B22.25), partial (25%)
Length = 664
Score = 35.8 bits (81), Expect = 0.061
Identities = 34/112 (30%), Positives = 43/112 (38%), Gaps = 4/112 (3%)
Frame = -1
Query: 232 PYPSTGPKKLILPIHT*PKSPAHQHKLPLSRGLYWNGPQES*PTTTTE*KSRGPTPFYPT 291
P+ PK + T*P +P H P G Q P T R P+P P
Sbjct: 361 PHSPRAPKWAVPNQPT*PTAPTSPH--PQLSG------QPPPPPPRTPAPPRNPSPDPP- 209
Query: 292 SCFELIPKTSYS----HHDPTPLEEPPQNHPKKPNPIKLLRRVPSRRRRRTP 339
P+TS+S HH P+ + P +HP PI P R RTP
Sbjct: 208 ------PQTSHSPTRFHHRPSTARDTPNSHPSPSAPIS-----PPPPRTRTP 86
>AV780221
Length = 461
Score = 35.4 bits (80), Expect = 0.080
Identities = 29/116 (25%), Positives = 51/116 (43%), Gaps = 1/116 (0%)
Frame = +3
Query: 1221 LIKSVIQSIPTYAMAVTPFPKG-FCKELDSLVARFWWSNNKKSRGIHLKDWHSLTCSKKE 1279
LI+SV+ ++P + + P G F +++ F+ + + I W ++ K E
Sbjct: 9 LIRSVLIALPLFHLPFFKLPTGGFYTNVNN**DPFFGREVEGGKKIAWVKWSTVCRPKDE 188
Query: 1280 GGMGFREFHLQNLAHMTKQAWRIHNTPHTLVAQVMKALYFPDSSFLDATPKPGDSW 1335
GG+G + L N A + K WR+ L +V+ Y +A P+ SW
Sbjct: 189 GGLGVQNLGLFNKALLGKWRWRMLKERDGLWYKVLLIKY------KNAIPQSASSW 338
>TC10496
Length = 562
Score = 34.7 bits (78), Expect = 0.14
Identities = 15/38 (39%), Positives = 22/38 (57%)
Frame = +3
Query: 429 WNCRGVGASTTARELKEINSKQKPSILFLMETRARRNK 466
WN RG+G+ST K+I K KP + L E++ N+
Sbjct: 444 WNVRGLGSSTKREAAKKILLKIKPEVCLLQESKLDENR 557
>BP038355
Length = 549
Score = 26.6 bits (57), Expect(2) = 0.19
Identities = 14/35 (40%), Positives = 21/35 (60%)
Frame = +1
Query: 1251 VARFWWSNNKKSRGIHLKDWHSLTCSKKEGGMGFR 1285
+AR W K RG H + H+L+ ++KE G+G R
Sbjct: 97 LARLW*YG--KDRGTH*VNLHTLSANRKEIGIGSR 195
Score = 26.2 bits (56), Expect(2) = 0.19
Identities = 14/28 (50%), Positives = 18/28 (64%)
Frame = +3
Query: 1216 AGKEVLIKSVIQSIPTYAMAVTPFPKGF 1243
+GK VL +S S PTYA ++ FPK F
Sbjct: 3 SGKGVL*RSKF-STPTYAFSIRKFPKHF 83
>BP067424
Length = 473
Score = 31.2 bits (69), Expect = 1.5
Identities = 21/50 (42%), Positives = 25/50 (50%), Gaps = 8/50 (16%)
Frame = +3
Query: 300 TSYSHHDPTPLEEPPQNHPKK-----PNPIKLLRRVPSRR---RRRTPKK 341
T+ SHH L PP P + P P +LLRR RR RRRTP +
Sbjct: 309 TAPSHHHEKLLHRPPLQPPHRQPHRLPWPYRLLRRGIHRRASLRRRTPPR 458
>TC13129 similar to UP|RIR2_TOBAC (P49730) Ribonucleoside-diphosphate
reductase small chain (Ribonucleotide reductase) (R2
subunit) , partial (46%)
Length = 482
Score = 30.8 bits (68), Expect = 2.0
Identities = 22/68 (32%), Positives = 31/68 (45%)
Frame = +3
Query: 267 NGPQES*PTTTTE*KSRGPTPFYPTSCFELIPKTSYSHHDPTPLEEPPQNHPKKPNPIKL 326
+ P P+T + +SR TP SC + +T PL+ P + P KP+P
Sbjct: 297 SNPPRHAPSTASRSRSRTSTPRCTASC*KPTSRT--------PLKNPTSSAPWKPSPAS- 449
Query: 327 LRRVPSRR 334
RR PS R
Sbjct: 450 -RRKPSGR 470
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.331 0.143 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 37,500,427
Number of Sequences: 28460
Number of extensions: 626470
Number of successful extensions: 4377
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 4203
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4353
length of query: 1797
length of database: 4,897,600
effective HSP length: 103
effective length of query: 1694
effective length of database: 1,966,220
effective search space: 3330776680
effective search space used: 3330776680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0304.11


